Cross-validating mixed-effects models
John Fox and Georges Monette
2025-09-24
Source:vignettes/cv-mixed.Rmd
cv-mixed.RmdThe fundamental analogy for cross-validation is to the collection of new data. That is, predicting the response in each fold from the model fit to data in the other folds is like using the model fit to all of the data to predict the response for new cases from the values of the predictors for those new cases. As we explained in the introductory vignette on cross-validating regression models, the application of this idea to independently sampled cases is straightforward—simply partition the data into random folds of equal size and leave each fold out in turn, or, in the case of LOO CV, simply omit each case in turn.
In contrast, mixed-effects models are fit to dependent data, in which cases as clustered, such as hierarchical data, where the clusters comprise higher-level units (e.g., students clustered in schools), or longitudinal data, where the clusters are individuals and the cases repeated observations on the individuals over time.1
We can think of two approaches to applying cross-validation to clustered data:2
Treat CV as analogous to predicting the response for one or more cases in a newly observed cluster. In this instance, the folds comprise one or more whole clusters; we refit the model with all of the cases in clusters in the current fold removed; and then we predict the response for the cases in clusters in the current fold. These predictions are based only on fixed effects because the random effects for the omitted clusters are presumably unknown, as they would be for data on cases in newly observed clusters.
Treat CV as analogous to predicting the response for a newly observed case in an existing cluster. In this instance, the folds comprise one or more individual cases, and the predictions can use both the fixed and random effects.
Example: The High-School and Beyond data
Following their use by Raudenbush & Bryk (2002), data from the 1982 High School and Beyond (HSB) survey have become a staple of the literature on mixed-effects models. The HSB data are used by Fox & Weisberg (2019, sec. 7.2.2) to illustrate the application of linear mixed models to hierarchical data, and we’ll closely follow their example here.
The HSB data are included in the MathAchieve and
MathAchSchool data sets in the nlme
package (Pinheiro & Bates, 2000).
MathAchieve includes individual-level data on 7185 students
in 160 high schools, and MathAchSchool includes
school-level data:
data("MathAchieve", package = "nlme")
dim(MathAchieve)
#> [1] 7185 6
head(MathAchieve, 3)
#> Grouped Data: MathAch ~ SES | School
#> School Minority Sex SES MathAch MEANSES
#> 1 1224 No Female -1.528 5.876 -0.428
#> 2 1224 No Female -0.588 19.708 -0.428
#> 3 1224 No Male -0.528 20.349 -0.428
tail(MathAchieve, 3)
#> Grouped Data: MathAch ~ SES | School
#> School Minority Sex SES MathAch MEANSES
#> 7183 9586 No Female 1.332 19.641 0.627
#> 7184 9586 No Female -0.008 16.241 0.627
#> 7185 9586 No Female 0.792 22.733 0.627
data("MathAchSchool", package = "nlme")
dim(MathAchSchool)
#> [1] 160 7
head(MathAchSchool, 2)
#> School Size Sector PRACAD DISCLIM HIMINTY MEANSES
#> 1224 1224 842 Public 0.35 1.597 0 -0.428
#> 1288 1288 1855 Public 0.27 0.174 0 0.128
tail(MathAchSchool, 2)
#> School Size Sector PRACAD DISCLIM HIMINTY MEANSES
#> 9550 9550 1532 Public 0.45 0.791 0 0.059
#> 9586 9586 262 Catholic 1.00 -2.416 0 0.627The first few students are in school number 1224 and the last few in school 9586.
We’ll use only the School, SES (students’
socioeconomic status), and MathAch (their score on a
standardized math-achievement test) variables in the
MathAchieve data set, and Sector
("Catholic" or "Public") in the
MathAchSchool data set.
Some data-management is required before fitting a mixed-effects model to the HSB data, for which we use the dplyr package (Wickham, François, Henry, Müller, & Vaughan, 2023):
library("dplyr")
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
MathAchieve %>% group_by(School) %>%
summarize(mean.ses = mean(SES)) -> Temp
Temp <- merge(MathAchSchool, Temp, by = "School")
HSB <- merge(Temp[, c("School", "Sector", "mean.ses")],
MathAchieve[, c("School", "SES", "MathAch")], by = "School")
names(HSB) <- tolower(names(HSB))
HSB$cses <- with(HSB, ses - mean.ses)In the process, we created two new school-level variables:
meanses, which is the average SES for students in each
school; and cses, which is school-average SES centered at
its mean. For details, see Fox & Weisberg
(2019, sec. 7.2.2).
Still following Fox and Weisberg, we proceed to use the
lmer() function in the lme4 package (Bates, Mächler, Bolker, & Walker, 2015) to
fit a mixed model for math achievement to the HSB data:
library("lme4")
hsb.lmer <- lmer(mathach ~ mean.ses * cses + sector * cses
+ (cses | school), data = HSB)
summary(hsb.lmer, correlation = FALSE)
#> Linear mixed model fit by REML ['lmerMod']
#> Formula: mathach ~ mean.ses * cses + sector * cses + (cses | school)
#> Data: HSB
#>
#> REML criterion at convergence: 46504
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -3.159 -0.723 0.017 0.754 2.958
#>
#> Random effects:
#> Groups Name Variance Std.Dev. Corr
#> school (Intercept) 2.380 1.543
#> cses 0.101 0.318 0.39
#> Residual 36.721 6.060
#> Number of obs: 7185, groups: school, 160
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> (Intercept) 12.128 0.199 60.86
#> mean.ses 5.333 0.369 14.45
#> cses 2.945 0.156 18.93
#> sectorCatholic 1.227 0.306 4.00
#> mean.ses:cses 1.039 0.299 3.48
#> cses:sectorCatholic -1.643 0.240 -6.85We can then cross-validate at the cluster (i.e., school) level,
library("cv")
summary(cv(hsb.lmer,
k = 10,
clusterVariables = "school",
seed = 5240))
#> R RNG seed set to 5240
#> 10-Fold Cross Validation based on 160 {school} clusters
#> criterion: mse
#> cross-validation criterion = 39.157
#> bias-adjusted cross-validation criterion = 39.148
#> 95% CI for bias-adjusted CV criterion = (38.066, 40.231)
#> full-sample criterion = 39.006or at the case (i.e., student) level,
summary(cv(hsb.lmer, seed = 1575))
#> R RNG seed set to 1575
#> Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
#> Model failed to converge with max|grad| = 0.00587207 (tol = 0.002, component 1)
#> boundary (singular) fit: see help('isSingular')
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 37.445
#> bias-adjusted cross-validation criterion = 37.338
#> 95% CI for bias-adjusted CV criterion = (36.288, 38.388)
#> full-sample criterion = 36.068For cluster-level CV, the clusterVariables argument
tells cv() how the clusters are defined. Were there more
than one clustering variable, say classes within schools, these would be
provided as a character vector of variable names:
clusterVariables = c("school", "class"). For cluster-level
CV, the default is k = "loo", that is, leave one cluster
out at a time; we instead specify k = 10 folds of clusters,
each fold therefore comprising
schools.
If the clusterVariables argument is omitted, then
case-level CV is employed, with k = 10 folds as the
default, here each with
students. Notice that one of the 10 models refit with a fold removed
failed to converge. Convergence problems are common in mixed-effects
modeling. The apparent issue here is that an estimated variance
component is close to or equal to 0, which is at a boundary of the
parameter space. That shouldn’t disqualify the fitted model for the kind
of prediction required for cross-validation.
There is also a cv() method for linear mixed models fit
by the lme() function in the nlme package,
and the arguments for cv() in this case are the same as for
a model fit by lmer() or glmer(). We
illustrate with the mixed model fit to the HSB data:
library("nlme")
#>
#> Attaching package: 'nlme'
#> The following object is masked from 'package:dplyr':
#>
#> collapse
#> The following object is masked from 'package:lme4':
#>
#> lmList
hsb.lme <- lme(
mathach ~ mean.ses * cses + sector * cses,
random = ~ cses | school,
data = HSB,
control = list(opt = "optim")
)
summary(hsb.lme)
#> Linear mixed-effects model fit by REML
#> Data: HSB
#> AIC BIC logLik
#> 46525 46594 -23252
#>
#> Random effects:
#> Formula: ~cses | school
#> Structure: General positive-definite, Log-Cholesky parametrization
#> StdDev Corr
#> (Intercept) 1.541177 (Intr)
#> cses 0.018174 0.006
#> Residual 6.063492
#>
#> Fixed effects: mathach ~ mean.ses * cses + sector * cses
#> Value Std.Error DF t-value p-value
#> (Intercept) 12.1282 0.19920 7022 60.886 0e+00
#> mean.ses 5.3367 0.36898 157 14.463 0e+00
#> cses 2.9421 0.15122 7022 19.456 0e+00
#> sectorCatholic 1.2245 0.30611 157 4.000 1e-04
#> mean.ses:cses 1.0444 0.29107 7022 3.588 3e-04
#> cses:sectorCatholic -1.6421 0.23312 7022 -7.044 0e+00
#> Correlation:
#> (Intr) men.ss cses sctrCt mn.ss:
#> mean.ses 0.256
#> cses 0.000 0.000
#> sectorCatholic -0.699 -0.356 0.000
#> mean.ses:cses 0.000 0.000 0.295 0.000
#> cses:sectorCatholic 0.000 0.000 -0.696 0.000 -0.351
#>
#> Standardized Within-Group Residuals:
#> Min Q1 Med Q3 Max
#> -3.170106 -0.724877 0.014892 0.754263 2.965498
#>
#> Number of Observations: 7185
#> Number of Groups: 160
summary(cv(hsb.lme,
k = 10,
clusterVariables = "school",
seed = 5240))
#> R RNG seed set to 5240
#> 10-Fold Cross Validation based on 160 {school} clusters
#> criterion: mse
#> cross-validation criterion = 39.157
#> bias-adjusted cross-validation criterion = 39.149
#> 95% CI for bias-adjusted CV criterion = (38.066, 40.232)
#> full-sample criterion = 39.006
summary(cv(hsb.lme, seed = 1575))
#> R RNG seed set to 1575
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 37.442
#> bias-adjusted cross-validation criterion = 37.402
#> 95% CI for bias-adjusted CV criterion = (36.351, 38.453)
#> full-sample criterion = 36.147We used the same random-number generator seeds as in the previous
example cross-validating the model fit by lmer(), and so
the same folds are employed in both cases.3 The estimated
covariance components and fixed effects in the summary output differ
slightly between the lmer() and lme()
solutions, although both functions seek to maximize the REML criterion.
This is, of course, to be expected when different algorithms are used
for numerical optimization. To the precision reported, the cluster-level
CV results for the lmer() and lme() models are
identical, while the case-level CV results are very similar but not
identical.
Example: Contrasting cluster-based and case-based CV
We introduce four artificial data sets that exemplify aspects of cross-validation particular to hierarchical models. Using these data sets, we show that model comparisons employing cluster-based and those employing case-based cross-validation may not agree on a “best” model. Furthermore, commonly used measures of fit, such as mean-squared error, do not necessarily become smaller as models become larger, even when the models are nested, and even when the measure of fit is computed for the whole data set.
The four datasets differ in the relative magnitude of between-cluster variance compared with within-cluster variance. They serve to illustrate how fitting mixed models, and, consequently, the cross-validation of mixed models, is sensitive to relative variance, which determines the degree of shrinkage of within-cluster estimates of effects towards between-cluster estimates.
For these analyses, we will use the glmmTMB() function
in the glmmTMB package (Brooks
et al., 2017) because, in our experience, it is more likely to
converge than functions in the nlme and the
lme4 packages for models with low between-cluster
variance.
Consider a researcher studying the effect of the dosage of a drug on the severity of symptoms for a hypothetical disease. The researcher has longitudinal data on 20 patients, each of whom was observed on five occasions in which patients received different dosages of the drug. The data are observational, with dosages prescribed by the patients’ physicians, so that patients who were more severely affected by the disease received higher dosages of the drug.
Our four contrived data sets (see below) illustrate possible results
for data obtained in such a scenario. The relative configuration of
dosages x and symptoms y are identical within
patients in each of the four datasets. Within patients, higher dosages
are generally associated with a reduction in symptoms.
Between patients, however, higher dosages are associated with higher levels of symptoms. A plausible mechanism is a reversal of causality: Within patients, higher dosages alleviate symptoms, but between patients higher morbidity causes the prescription of higher dosages.
The four data sets differ in the between-patient variance of patient centroids from a common between-patients regression line. The data sets exhibit a progression from low to high variance around the common regression line.
We start by generating a data set to serve as a common template for the four sample data sets. We then apply different multipliers to the between-patient variation using parameters consistent with the description above to highlight the issues that arise in cross-validating mixed-effects models:4
# Parameters:
Nb <- 20 # number of patients
Nw <- 5 # number of occasions for each patient
Bb <- 1.0 # between-patient regression coefficient on patient means
Bw <- -0.5 # within-patient effect of x
SD_between <- c(0, 5, 6, 8) # SD between patients
SD_within <- rep(2.5, length(SD_between)) # SD within patients
Nv <- length(SD_within) # number of variance profiles
SD_ratio <- paste0('SD ratio = ', SD_between,' / ',SD_within)
SD_ratio <- factor(SD_ratio, levels = SD_ratio)
set.seed(833885)
Data_template <- expand.grid(patient = 1:Nb, obs = 1:Nw) |>
within({
xw <- seq(-2, 2, length.out = Nw)[obs]
x <- patient + xw
xm <- ave(x, patient) # within-patient mean
# Scaled random error within each SD_ratio_i group
re_std <- scale(resid(lm(rnorm(Nb*Nw) ~ x)))
re_between <- ave(re_std, patient)
re_within <- re_std - re_between
re_between <- scale(re_between)/sqrt(Nw)
re_within <- scale(re_within)
})
Data <- do.call(
rbind,
lapply(
1:Nv,
function(i) {
cbind(Data_template, SD_ratio_i = i)
}
)
)
Data <- within(
Data,
{
SD_within_ <- SD_within[SD_ratio_i]
SD_between_ <- SD_between[SD_ratio_i]
SD_ratio <- SD_ratio[SD_ratio_i]
y <- 10 +
Bb * xm + # contextual effect
Bw * (x - xm) + # within-patient effect
SD_within_ * re_within + # within patient random effect
SD_between_ * re_between # adjustment to between patient random effect
}
)Here is a scatterplot of the data sets showing estimated 50% concentration ellipses for each cluster:5
library("lattice")
library("latticeExtra")
plot <- xyplot(y ~ x | SD_ratio, data = Data, group = patient,
layout = c(Nv, 1),
par.settings = list(superpose.symbol = list(pch = 1, cex = 0.7))) +
layer(panel.ellipse(..., center.pch = 16, center.cex = 1.5,
level = 0.5),
panel.abline(a = 10, b = 1))
plot # display graph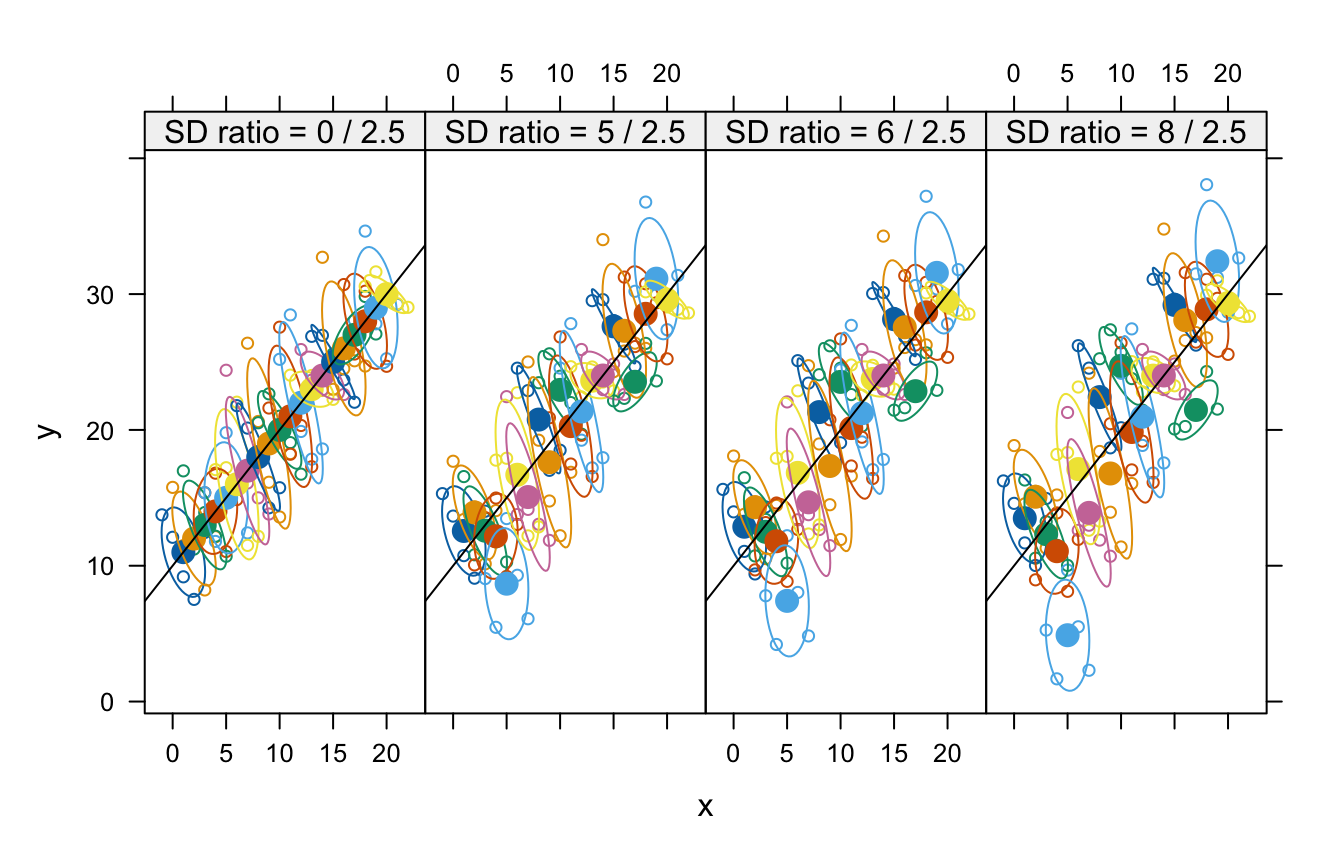
Data sets showing identical within-patient configurations with increasing between-patient variance. The labels above each panel show the between-patient SD/within-patient SD. The ellipses are estimated 50% concentration ellipses for each patient. The population between-patient regression line, , is shown in each panel.
The population from which these datasets is generated has a
between-patient effect of dosage of 1 and a within-patient effect of
.
The researcher may attempt to obtain an estimate of the within-patient
effect of dosage through the use of mixed models with a random
intercept. We will illustrate how this approach results in estimates
that are highly sensitive to the relative variance at the two levels of
the mixed model by considering four models, all of which include a
random intercept. The first three models have sequentially nested fixed
effects: (1)~ 1, intercept only; (2)~ 1 + x,
intercept and effect of x; and (3)
~ 1 + x + xm, intercept, effect of x, and a
contextual variable, xm, consisting of the within-patient
mean of x. The fourth model, ~ 1 + I(x - xm),
uses an intercept and the centered-within-group variable
x - xm. We thus fit four models to four datasets for a
total of 16 models.
model.formulas <- c(
' ~ 1' = y ~ 1 + (1 | patient),
'~ 1 + x' = y ~ 1 + x + (1 | patient),
'~ 1 + x + xm' = y ~ 1 + x + xm + (1 | patient),
'~ 1 + I(x - xm)' = y ~ 1 + I(x - xm) + (1 | patient)
)
fits <- lapply(split(Data, ~ SD_ratio),
function(d) {
lapply(model.formulas, function(form) {
glmmTMB(form, data = d)
})
})We proceed to obtain predictions from each model based on fixed effects alone, as would be used for cross-validation based on clusters (i.e., patients), and for fixed and random effects—so-called best linear unbiased predictions or “BLUPs”—as would be used for cross-validation based on cases (i.e., occasions within patients):
# predicted fixed and random effects:
pred.BLUPs <- lapply(fits, lapply, predict)
# predicted fixed effects:
pred.fixed <- lapply(fits, lapply, predict, re.form = ~0) We then prepare the data for plotting:
Dataf <- lapply(split(Data, ~ SD_ratio),
function(d) {
lapply(names(model.formulas),
function(form) cbind(d, formula = form))
}) |>
lapply(function(dlist) do.call(rbind, dlist)) |>
do.call(rbind, args = _)
Dataf <- within(
Dataf,
{
pred.fixed <- unlist(pred.fixed)
pred.BLUPs <- unlist(pred.BLUPs)
panel <- factor(formula, levels = c(names(model.formulas), 'data'))
}
)
Data$panel <- factor('data', levels = c(names(model.formulas), 'data'))The fixed-effects predictions from these models are shown in the following graph:
{
xyplot(y ~ x |SD_ratio * panel, Data,
groups = patient, type = 'n',
par.strip.text = list(cex = 0.7),
drop.unused.levels = FALSE) +
glayer(panel.ellipse(..., center.pch = 16, center.cex = 0.5,
level = 0.5),
panel.abline(a = 10, b = 1)) +
xyplot(pred.fixed ~ x |SD_ratio * panel, Dataf, type = 'l',
groups = patient,
drop.unused.levels = F,
ylab = 'fixed-effect predictions')
}|>
useOuterStrips() |> print()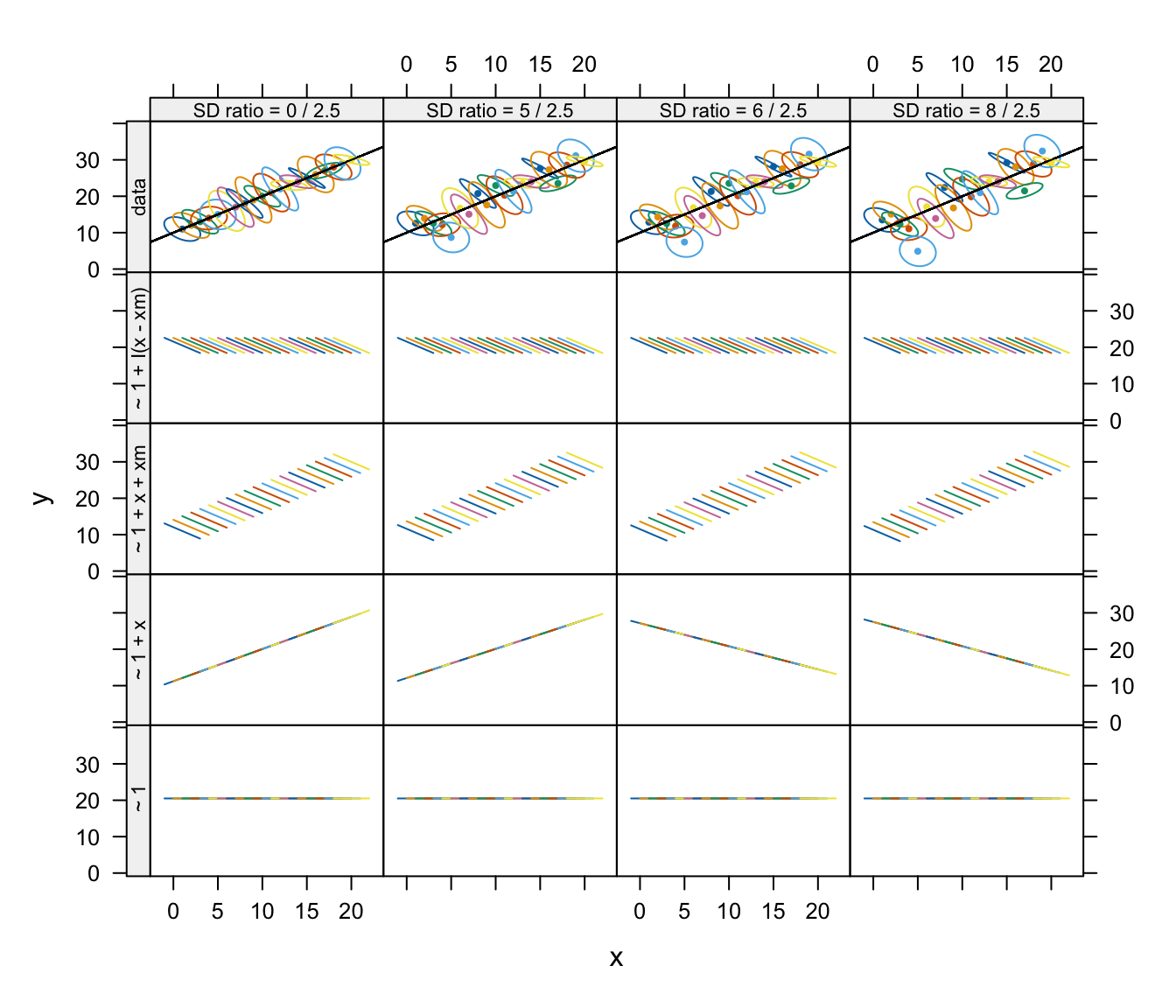
Fixed-effect predictions using each model applied to data sets with varying between- and within-patient variance ratios. The top row shows summaries of the within-patient data using estimated 50% concentration ellipses for each patient.
The BLUPs from these models are shown in the following graph:
{
xyplot(y ~ x | SD_ratio * panel, Data,
groups = patient, type = 'n',
drop.unused.levels = F,
par.strip.text = list(cex = 0.7),
ylab = 'fixed- and random-effect predictions (BLUPS)') +
glayer(panel.ellipse(..., center.pch = 16, center.cex = 0.5,
level = 0.5),
panel.abline(a = 10, b = 1)) +
xyplot(pred.BLUPs ~ x | SD_ratio * panel, Dataf, type = 'l',
groups = patient,
drop.unused.levels = F)
}|>
useOuterStrips() |> print()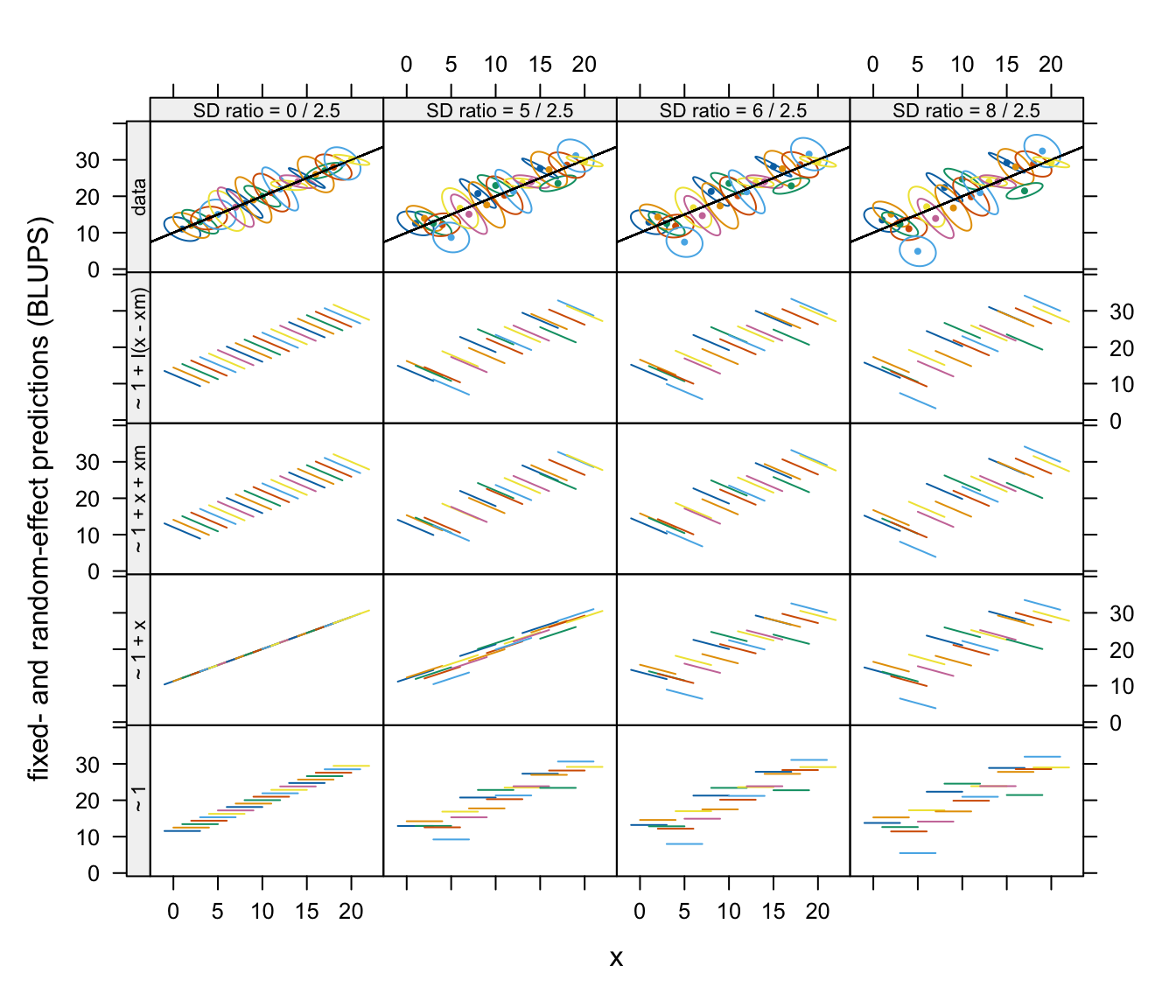
Fixed- and random-effect predictions (BLUPs) using each model applied to data sets with varying between- and within-patient variance ratios. The top row shows summaries of the within-patient data using estimated 50% concentration ellipses for each patient.
Data sets with relatively low between-patient variance result in
strong shrinkage of fixed-effects predictions and also of BLUPs towards
the between-patient relationship between y and
x in the model with an intercept and x as
fixed-effect predictors, ~ 1 + x. The inclusion of a
contextual variable in the model corrects this problem.
Although the BLUPs fit the observed data more closely than
predictions based on fixed effects alone, the slopes of within-patient
BLUPs do not conform with the within-patient slopes for the
~ 1 + x model in the two datasets with the smallest
between-patient variances.
For data with a small between-patient variance, fixed-effects
predictions for the ~ 1 + x model have a slope that is
close to the between-patient slope but provide better overall
predictions than the fixed-effect predictions for datasets with larger
between-subject variance. With data whose between-patient variance is
relatively large, predictions based on the model with a common intercept
and slope for all clusters, are very poor—indeed, much worse than the
fixed-effects-only predictions based on the simpler random-intercept
model.
We therefore anticipate (and show later in this section) that
case-based cross-validation may prefer the intercept-only model,
~ 1 to the larger ~ 1 + x model when the
between-cluster variance is relatively small, but that cluster-based
cross-validation will prefer the latter to the former.
We will discover that case-based cross-validation prefers the
~ 1 + x model to the ~ 1 model for the ‘5 /
2.5’ dataset, but cluster-based cross-validation prefers the latter
model to the former. The situation is entirely reversed with the ‘8 /
2.5’ dataset.
The third model, ~ 1 + x + xm, includes a contextual
effect of x—that is, the cluster mean xm—along
with x and the intercept in the fixed-effect part of the
model, along with a random intercept. This model is equivalent to
fitting y ~ I(x - xm) + xm + (1 | patient), which is the
model that generated the data. The fit of the mixed model
~ 1 + x + xm is consequently similar to that of a
fixed-effects only model with x and a categorical predictor
for individual patients (i.e., y ~ factor(patient) + x,
treating patients as a factor, and not shown here).
We next carry out case-based cross-validation, which, as we have
explained, is based on both fixed and predicted random effects (i.e.,
BLUPs), and cluster-based cross-validation, which is based on fixed
effects only. In order to reduce between-model random variability in
comparisons of models on the same dataset, we apply cv() to
the list of models created by the models() function
(introduced previously), performing cross-validation with the same folds
for each model.
model_lists <- lapply(fits, function(fitlist) do.call(models, fitlist))
cvs_cases <-
lapply(1:Nv,
function(i){
cv(model_lists[[i]], k = 10,
data = split(Data, ~ SD_ratio)[[i]])
})
#> R RNG seed set to 982470
#> R RNG seed set to 903914
#> R RNG seed set to 246168
#> R RNG seed set to 948407
cvs_clusters <-
lapply(1:Nv,
function(i){
cv(model_lists[[i]], k = 10,
data = split(Data, ~SD_ratio)[[i]],
clusterVariables = 'patient')
})
#> R RNG seed set to 382198
#> R RNG seed set to 533366
#> R RNG seed set to 637459
#> R RNG seed set to 542289For a given dataset, we can plot the results for a list of models
using the plot method for "cvList" objects. For example
plot(cvs_clusters[[2]], main="Comparison of Fixed Effects")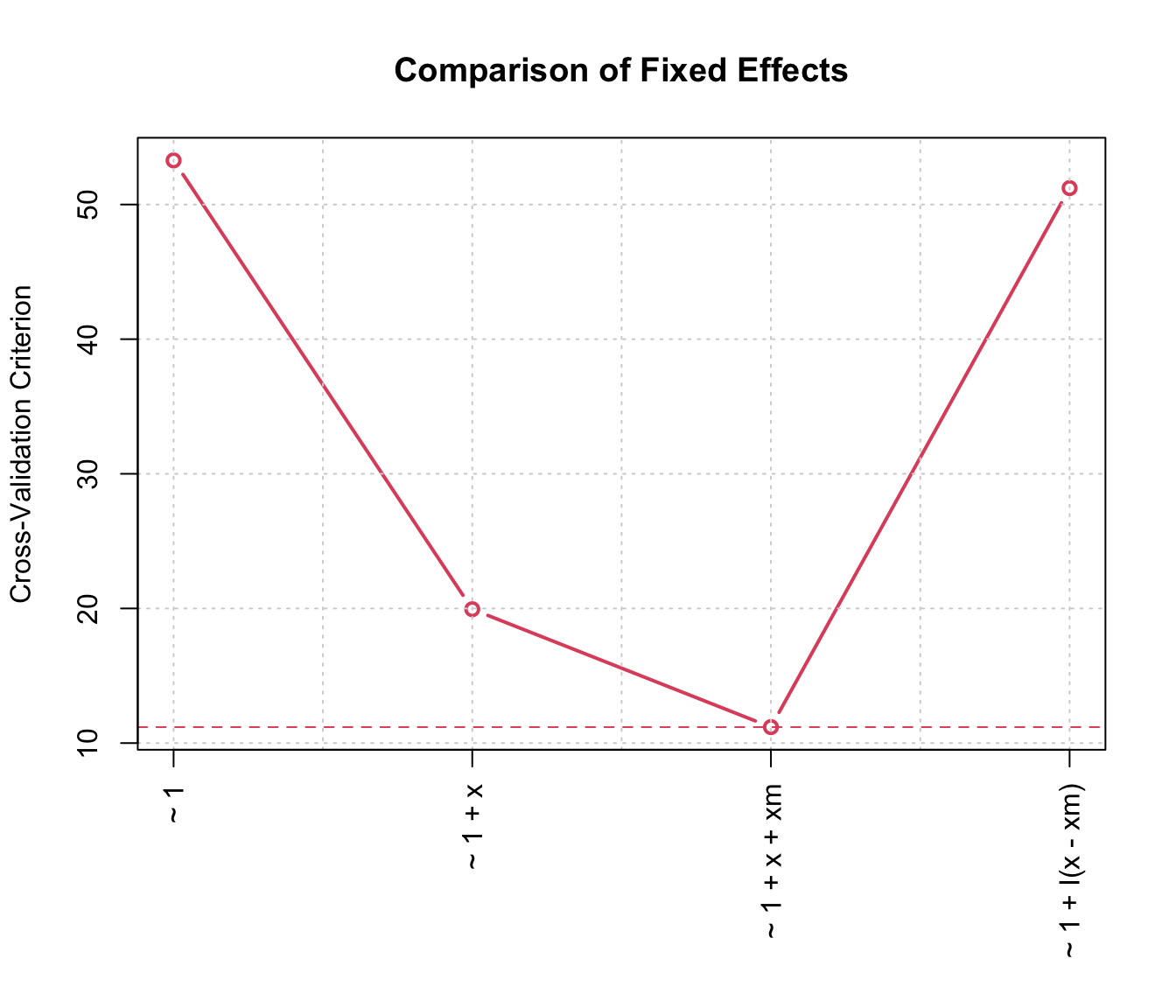
10-fold cluster-based cross-validation comparing random intercept mixed models with varying fixed effects.
We assemble the results for all datasets to show them in a common figure.
names(cvs_clusters) <- names(cvs_cases) <- SD_ratio
dsummary <- expand.grid(SD_ratio_i = names(cvs_cases), model = names(cvs_cases[[1]]))
dsummary$cases <-
sapply(1:nrow(dsummary), function(i){
with(dsummary[i,], cvs_cases[[SD_ratio_i]][[model]][['CV crit']])
})
dsummary$clusters <-
sapply(1:nrow(dsummary), function(i){
with(dsummary[i,], cvs_clusters[[SD_ratio_i]][[model]][['CV crit']])
})
xyplot(cases + clusters ~ model|SD_ratio_i, dsummary,
auto.key = list(space = 'top', reverse.rows = T, columns = 2), type = 'b',
xlab = "Fixed Effects",
ylab = 'CV criterion (MSE)',
layout= c(Nv,1),
par.settings =
list(superpose.line=list(lty = c(2, 3), lwd = 3),
superpose.symbol=list(pch = 15:16, cex = 1.5)),
scales = list(y = list(log = TRUE), x = list(alternating = F, rot = 60))) |> print()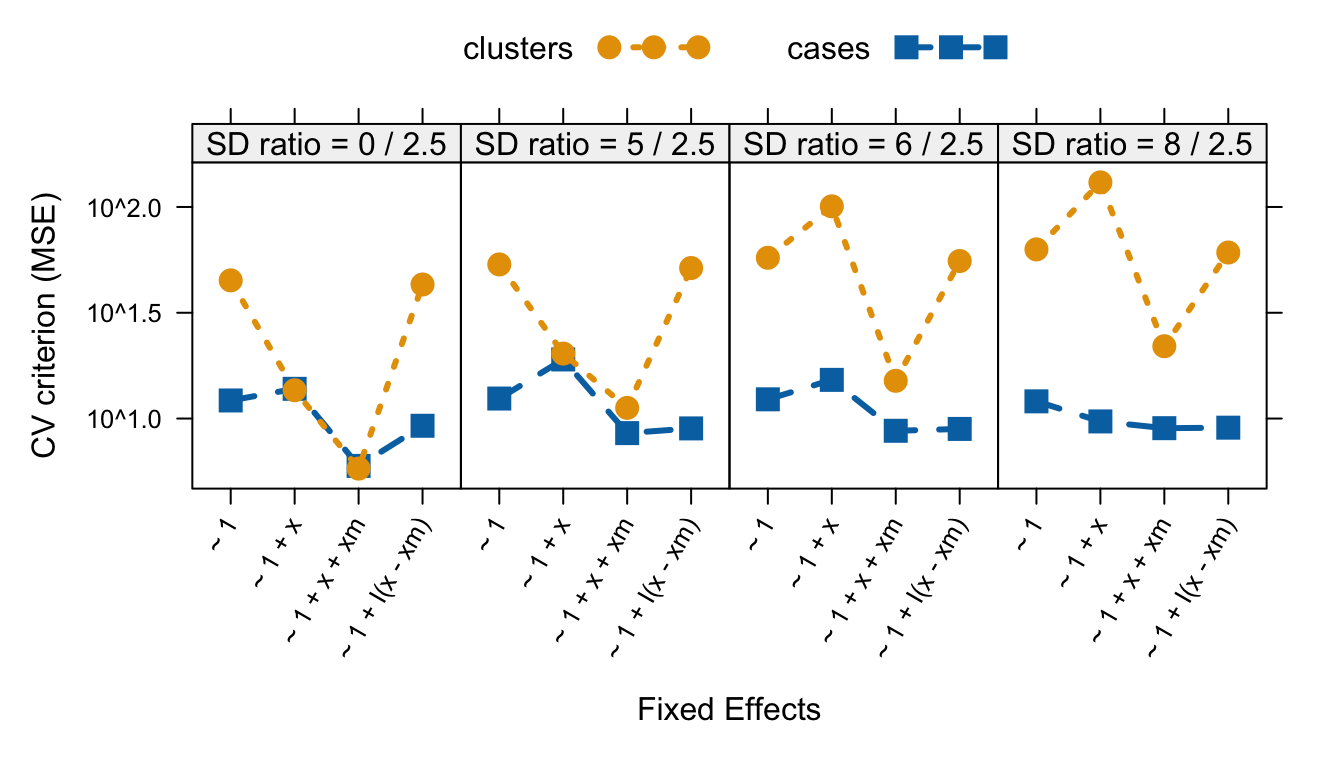
10-fold cluster- and case-based cross-validation comparing mixed models with random intercepts and various fixed effects.
In summary, when between-cluster variance is relatively large, the
model ~ 1 + x, with x alone and without the
contextual mean of x, is assessed as fitting very poorly by
cluster-based CV, but relatively much better by case-based CV. In all
our examples, the model ~ 1 + x + xm, which includes both
x and its contextual mean, produces better results using
both cluster-based and case-based CV. These conclusions are consistent
with our observations based on graphing predictions from the various
models, and they illustrate the desirability of assessing mixed-effect
models at different hierarchical levels.
Example: Crossed random effects
Crossed random effects arise when the structure of the data aren’t strictly hierarchical. Nevertheless, crossed and nested random effects can be handled in much the same manner, by refitting the mixed-effects model to the data with a fold of clusters or cases removed and using the refitted model to predict the response in the removed fold.
We’ll illustrate with data on pig growth, introduced by Diggle, Liang, & Zeger (1994, Table 3.1).
The data are in the Pigs data frame in the
cv package:
head(Pigs, 9)
#> id week weight
#> 1 1 1 24.0
#> 2 1 2 32.0
#> 3 1 3 39.0
#> 4 1 4 42.5
#> 5 1 5 48.0
#> 6 1 6 54.5
#> 7 1 7 61.0
#> 8 1 8 65.0
#> 9 1 9 72.0
head(xtabs( ~ id + week, data = Pigs), 3)
#> week
#> id 1 2 3 4 5 6 7 8 9
#> 1 1 1 1 1 1 1 1 1 1
#> 2 1 1 1 1 1 1 1 1 1
#> 3 1 1 1 1 1 1 1 1 1
tail(xtabs( ~ id + week, data = Pigs), 3)
#> week
#> id 1 2 3 4 5 6 7 8 9
#> 46 1 1 1 1 1 1 1 1 1
#> 47 1 1 1 1 1 1 1 1 1
#> 48 1 1 1 1 1 1 1 1 1Each of 48 pigs is observed weekly over a period of 9 weeks, with the
weight of the pig recorded in kg. The data are in “long” format, as is
appropriate for use with the lmer() function in the
lme4 package. The data are very regular, with no
missing cases.
The following graph, showing the growth trajectories of the pigs, is similar to Figure 3.1 in Diggle et al. (1994); we add an overall least-squares line and a loess smooth, which are nearly indistinguishable:
plot(weight ~ week, data = Pigs, type = "n")
for (i in unique(Pigs$id)) {
with(Pigs, lines(
x = 1:9,
y = Pigs[id == i, "weight"],
col = "gray"
))
}
abline(lm(weight ~ week, data = Pigs),
col = "blue",
lwd = 2)
lines(
with(Pigs, loess.smooth(week, weight, span = 0.5)),
col = "magenta",
lty = 2,
lwd = 2
)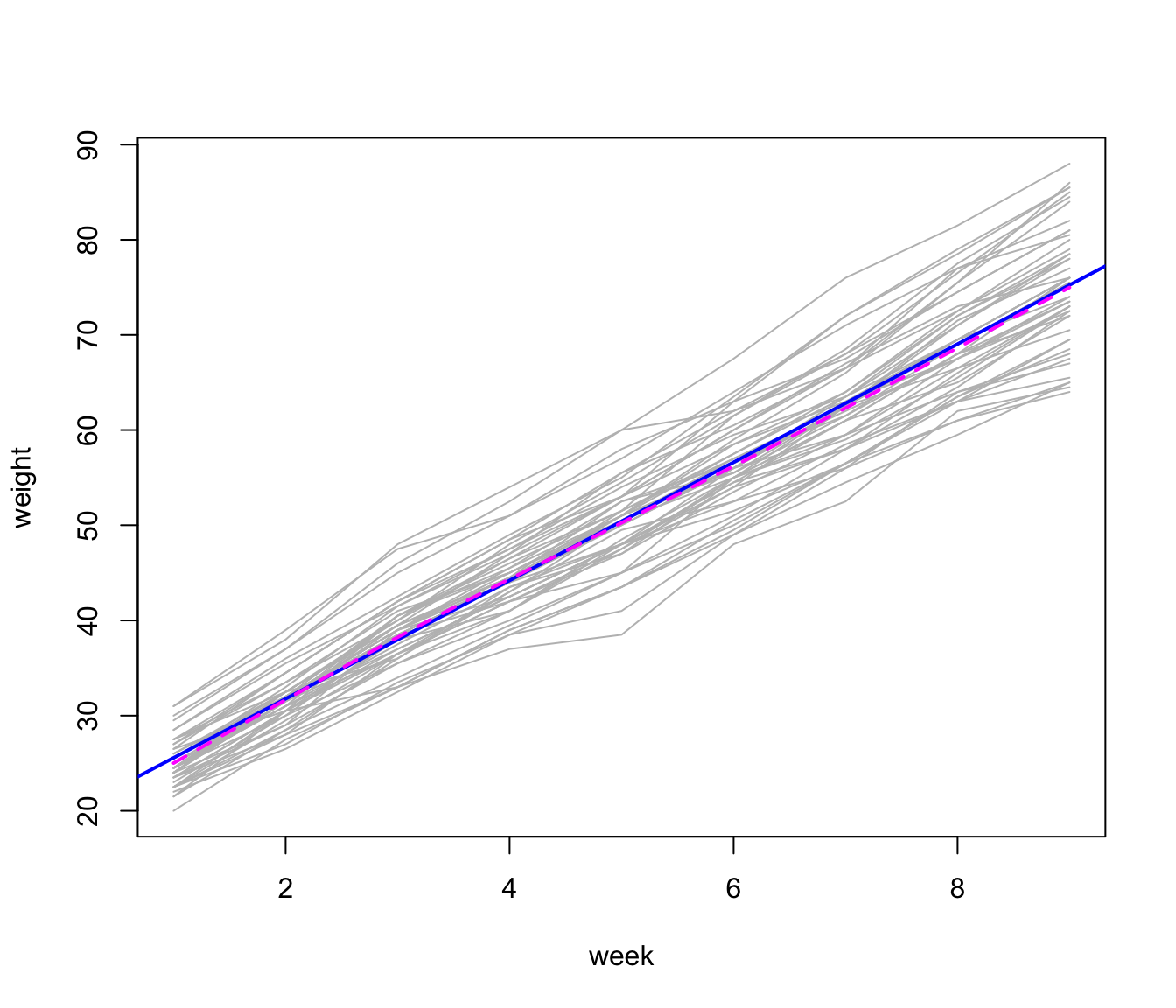
Growth trajectories for 48 pigs, with overall least-squares line (sold blue) and loess line (broken magenta).
The individual “growth curves” and the overall trend are generally linear, with some tendency for variability of pig weight to increase over weeks (a feature of the data that we ignore in the mixed model that we fit to the data below).
The Stata mixed-effects models manual proposes a
model with crossed random effects for the Pigs data (StataCorp LLC, 2023, p. 37):
[S]uppose that we wish to fit for the weeks and pigs and all independently. That is, we assume an overall population-average growth curve and a random pig-specific shift. In other words, the effect due to week, , is systematic to that week and common to all pigs. The rationale behind [this model] could be that, assuming that the pigs were measured contemporaneously, we might be concerned that week-specific random factors such as weather and feeding patterns had significant systematic effects on all pigs.
Although we might prefer an alternative model,6 we think that this is a reasonable specification.
The Stata manual fits the mixed model by maximum
likelihood (rather than REML), and we duplicate the results reported
there using lmer():
m.p <- lmer(
weight ~ week + (1 | id) + (1 | week),
data = Pigs,
REML = FALSE, # i.e., ML
control = lmerControl(optimizer = "bobyqa")
)
summary(m.p)
#> Linear mixed model fit by maximum likelihood ['lmerMod']
#> Formula: weight ~ week + (1 | id) + (1 | week)
#> Data: Pigs
#> Control: lmerControl(optimizer = "bobyqa")
#>
#> AIC BIC logLik -2*log(L) df.resid
#> 2037.6 2058.0 -1013.8 2027.6 427
#>
#> Scaled residuals:
#> Min 1Q Median 3Q Max
#> -3.775 -0.542 0.005 0.476 3.982
#>
#> Random effects:
#> Groups Name Variance Std.Dev.
#> id (Intercept) 14.836 3.852
#> week (Intercept) 0.085 0.292
#> Residual 4.297 2.073
#> Number of obs: 432, groups: id, 48; week, 9
#>
#> Fixed effects:
#> Estimate Std. Error t value
#> (Intercept) 19.3556 0.6334 30.6
#> week 6.2099 0.0539 115.1
#>
#> Correlation of Fixed Effects:
#> (Intr)
#> week -0.426We opt for the non-default "bobyqa" optimizer because it
provides more numerically stable results for subsequent cross-validation
in this example.
We can then cross-validate the model by omitting folds composed of
pigs, folds composed of weeks, or folds composed of pig-weeks (which in
the Pigs data set correspond to individual cases, using
only the fixed effects):
summary(cv(m.p, clusterVariables = "id"))
#> n-Fold Cross Validation based on 48 {id} clusters
#> criterion: mse
#> cross-validation criterion = 19.973
#> bias-adjusted cross-validation criterion = 19.965
#> 95% CI for bias-adjusted CV criterion = (17.125, 22.805)
#> full-sample criterion = 19.201
summary(cv(m.p, clusterVariables = "week"))
#> boundary (singular) fit: see help('isSingular')
#> n-Fold Cross Validation based on 9 {week} clusters
#> criterion: mse
#> cross-validation criterion = 19.312
#> bias-adjusted cross-validation criterion = 19.305
#> 95% CI for bias-adjusted CV criterion = (16.566, 22.044)
#> full-sample criterion = 19.201
summary(cv(
m.p,
clusterVariables = c("id", "week"),
k = 10,
seed = 8469))
#> R RNG seed set to 8469
#> 10-Fold Cross Validation based on 432 {id, week} clusters
#> criterion: mse
#> cross-validation criterion = 19.235
#> bias-adjusted cross-validation criterion = 19.233
#> 95% CI for bias-adjusted CV criterion = (16.493, 21.973)
#> full-sample criterion = 19.201We can also cross-validate the individual cases taking account of the random effects (employing the same 10 folds):
summary(cv(m.p, k = 10, seed = 8469))
#> R RNG seed set to 8469
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 5.1583
#> bias-adjusted cross-validation criterion = 5.0729
#> 95% CI for bias-adjusted CV criterion = (4.123, 6.0229)
#> full-sample criterion = 3.796Because these predictions are based on BLUPs, they are more accurate than the predictions based only on fixed effects.7 As well, the difference between the MSE computed for the model fit to the full data and the CV estimates of the MSE is greater here than for cluster-based predictions.