Cross-validating model selection
John Fox and Georges Monette
2025-09-24
Source:vignettes/cv-selection.Rmd
cv-selection.RmdAs Hastie, Tibshirani, & Friedman (2009, sec. 7.10.2: “The Wrong and Right Way to Do Cross-validation”) explain, if the whole data are used to select or fine-tune a statistical model, subsequent cross-validation of the model is intrinsically misleading, because the model is selected to fit the whole data, including the part of the data that remains when each fold is removed.
A preliminary example
The following example is similar in spirit to one employed by Hastie et al. (2009). Suppose that we randomly generate independent observations for a response variable variable , and independently sample observations for “predictors,” , each from . The response has nothing to do with the predictors and so the population linear-regression model has and all .
set.seed(24361) # for reproducibility
D <- data.frame(y = rnorm(1000, mean = 10),
X = matrix(rnorm(1000 * 100), 1000, 100))
head(D[, 1:6])
#> y X.1 X.2 X.3 X.4 X.5
#> 1 10.0316 -1.23886 -0.26487 -0.03539 -2.576973 0.811048
#> 2 9.6650 0.12287 -0.17744 0.37290 -0.935138 0.628673
#> 3 10.0232 -0.95052 -0.73487 -1.05978 0.882944 0.023918
#> 4 8.9910 1.13571 0.32411 0.11037 1.376303 -0.422114
#> 5 9.0712 1.49474 1.87538 0.10575 0.292140 -0.184568
#> 6 11.3493 -0.18453 -0.78037 -1.23804 -0.010949 0.691034Least-squares provides accurate estimates of the regression constant and the error variance for the “null model” including only the regression constant; moreover, the omnibus -test of the correct null hypothesis that all of the s are 0 for the “full model” with all 100 s is associated with a large -value:
m.full <- lm(y ~ ., data = D)
m.null <- lm(y ~ 1, data = D)
anova(m.null, m.full)
#> Analysis of Variance Table
#>
#> Model 1: y ~ 1
#> Model 2: y ~ X.1 + X.2 + X.3 + X.4 + X.5 + X.6 + X.7 + X.8 + X.9 + X.10 +
#> X.11 + X.12 + X.13 + X.14 + X.15 + X.16 + X.17 + X.18 + X.19 +
#> X.20 + X.21 + X.22 + X.23 + X.24 + X.25 + X.26 + X.27 + X.28 +
#> X.29 + X.30 + X.31 + X.32 + X.33 + X.34 + X.35 + X.36 + X.37 +
#> X.38 + X.39 + X.40 + X.41 + X.42 + X.43 + X.44 + X.45 + X.46 +
#> X.47 + X.48 + X.49 + X.50 + X.51 + X.52 + X.53 + X.54 + X.55 +
#> X.56 + X.57 + X.58 + X.59 + X.60 + X.61 + X.62 + X.63 + X.64 +
#> X.65 + X.66 + X.67 + X.68 + X.69 + X.70 + X.71 + X.72 + X.73 +
#> X.74 + X.75 + X.76 + X.77 + X.78 + X.79 + X.80 + X.81 + X.82 +
#> X.83 + X.84 + X.85 + X.86 + X.87 + X.88 + X.89 + X.90 + X.91 +
#> X.92 + X.93 + X.94 + X.95 + X.96 + X.97 + X.98 + X.99 + X.100
#> Res.Df RSS Df Sum of Sq F Pr(>F)
#> 1 999 974
#> 2 899 888 100 85.2 0.86 0.82
summary(m.null)
#>
#> Call:
#> lm(formula = y ~ 1, data = D)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -3.458 -0.681 0.019 0.636 2.935
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 9.9370 0.0312 318 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.987 on 999 degrees of freedomNext, using the stepAIC() function in the
MASS package (Venables &
Ripley, 2002), let us perform a forward stepwise regression to
select a “best” model, starting with the null model, and using AIC as
the model-selection criterion (see the help page for
stepAIC() for details):1
library("MASS") # for stepAIC()
m.select <- stepAIC(
m.null,
direction = "forward",
trace = FALSE,
scope = list(lower = ~ 1, upper = formula(m.full))
)
summary(m.select)
#>
#> Call:
#> lm(formula = y ~ X.99 + X.90 + X.87 + X.40 + X.65 + X.91 + X.53 +
#> X.45 + X.31 + X.56 + X.61 + X.60 + X.46 + X.35 + X.92, data = D)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -3.262 -0.645 0.024 0.641 3.118
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 9.9372 0.0310 320.80 <2e-16 ***
#> X.99 -0.0910 0.0308 -2.95 0.0032 **
#> X.90 -0.0820 0.0314 -2.62 0.0090 **
#> X.87 -0.0694 0.0311 -2.24 0.0256 *
#> X.40 -0.0476 0.0308 -1.55 0.1221
#> X.65 -0.0552 0.0315 -1.76 0.0795 .
#> X.91 0.0524 0.0308 1.70 0.0894 .
#> X.53 -0.0492 0.0305 -1.61 0.1067
#> X.45 0.0554 0.0318 1.74 0.0818 .
#> X.31 0.0452 0.0311 1.46 0.1457
#> X.56 0.0543 0.0327 1.66 0.0972 .
#> X.61 -0.0508 0.0317 -1.60 0.1091
#> X.60 -0.0513 0.0319 -1.61 0.1083
#> X.46 0.0516 0.0327 1.58 0.1153
#> X.35 0.0470 0.0315 1.49 0.1358
#> X.92 0.0443 0.0310 1.43 0.1533
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.973 on 984 degrees of freedom
#> Multiple R-squared: 0.0442, Adjusted R-squared: 0.0296
#> F-statistic: 3.03 on 15 and 984 DF, p-value: 8.34e-05
library("cv")
#> Loading required package: doParallel
#> Loading required package: foreach
#> Loading required package: iterators
#> Loading required package: parallel
mse(D$y, fitted(m.select))
#> [1] 0.93063
#> attr(,"casewise loss")
#> [1] "(y - yhat)^2"The resulting model has 15 predictors, a very modest , but a small -value for its omnibus -test (which, of course, is entirely spurious because the same data were used to select and test the model). The MSE for the selected model is smaller than the true error variance , as is the estimated error variance for the selected model, .
If we cross-validate the selected model, we also obtain an optimistic estimate of its predictive power (although the confidence interval for the bias-adjusted MSE includes 1):
library("cv")
summary(cv(m.select, seed = 2529))
#> R RNG seed set to 2529
#> 10-Fold Cross Validation
#> method: Woodbury
#> criterion: mse
#> cross-validation criterion = 0.95937
#> bias-adjusted cross-validation criterion = 0.95785
#> 95% CI for bias-adjusted CV criterion = (0.87661, 1.0391)
#> full-sample criterion = 0.93063The "function" method of cv() allows us to
cross-validate the whole model-selection procedure, where first argument
to cv() is a model-selection function capable of refitting
the model with a fold omitted and returning a CV criterion. The
selectStepAIC() function, in the cv
package and based on stepAIC(), is suitable for use with
cv():
cv.select <- cv(
selectStepAIC,
data = D,
seed = 3791,
working.model = m.null,
direction = "forward",
scope = list(lower = ~ 1, upper = formula(m.full))
)
#> R RNG seed set to 3791
summary(cv.select)
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 1.0687
#> bias-adjusted cross-validation criterion = 1.0612
#> 95% CI for bias-adjusted CV criterion = (0.97172, 1.1506)
#> full-sample criterion = 0.93063The other arguments to cv() are:
-
data, the data set to which the model is fit; -
seed, an optional seed for R’s pseudo-random-number generator; as forcv(), if the seed isn’t supplied by the user, a seed is randomly selected and saved; - additional arguments required by the model-selection function, here
the starting
working.modelargument, thedirectionof model selection, and thescopeof models considered (from the model with only a regression constant to the model with all 100 predictors).
By default, cv() performs 10-fold CV, and produces an
estimate of MSE for the model-selection procedure even larger
than the true error variance,
.
Also by default, when the number of folds is 10 or fewer,
cv() saves details data about the folds. In this example,
the compareFolds() function reveals that the variables
retained by the model-selection process in the several folds are quite
different:
compareFolds(cv.select)
#> CV criterion by folds:
#> fold.1 fold.2 fold.3 fold.4 fold.5 fold.6 fold.7 fold.8 fold.9 fold.10
#> 1.26782 1.12837 1.04682 1.31007 1.06899 0.87916 0.88380 0.95026 1.21070 0.94130
#>
#> Coefficients by folds:
#> (Intercept) X.87 X.90 X.99 X.91 X.54 X.53 X.56
#> Fold 1 9.9187 -0.0615 -0.0994 -0.0942 0.0512 0.0516
#> Fold 2 9.9451 -0.0745 -0.0899 -0.0614 0.0587 0.0673
#> Fold 3 9.9423 -0.0783 -0.0718 -0.0987 0.0601 0.0512
#> Fold 4 9.9410 -0.0860 -0.0831 -0.0867 0.0570 -0.0508
#> Fold 5 9.9421 -0.0659 -0.0849 -0.1004 0.0701 0.0511 -0.0487 0.0537
#> Fold 6 9.9633 -0.0733 -0.0874 -0.0960 0.0555 0.0629 -0.0478
#> Fold 7 9.9279 -0.0618 -0.0960 -0.0838 0.0533 -0.0464
#> Fold 8 9.9453 -0.0610 -0.0811 -0.0818 0.0497 -0.0612 0.0560
#> Fold 9 9.9173 -0.0663 -0.0894 -0.1100 0.0504 0.0524 0.0747
#> Fold 10 9.9449 -0.0745 -0.0906 -0.0891 0.0535 0.0482 -0.0583 0.0642
#> X.40 X.45 X.65 X.68 X.92 X.15 X.26 X.46 X.60
#> Fold 1 -0.0590 -0.0456 0.0658 0.0608
#> Fold 2 0.0607 0.0487
#> Fold 3 -0.0496 -0.0664 0.0494
#> Fold 4 -0.0597 0.0579 -0.0531 0.0519 -0.0566 -0.0519
#> Fold 5 0.0587 0.0527 -0.0603
#> Fold 6 -0.0596 0.0552 0.0474
#> Fold 7 0.0572 0.0595
#> Fold 8 0.0547 -0.0617 0.0453 0.0493 -0.0613 0.0591 0.0703 -0.0588
#> Fold 9 -0.0552 0.0573 -0.0635 0.0492 -0.0513 0.0484 -0.0507
#> Fold 10 -0.0558 0.0529 0.0710
#> X.61 X.8 X.28 X.29 X.31 X.35 X.70 X.89 X.17
#> Fold 1 -0.0490 0.0616 -0.0537 0.0638
#> Fold 2 0.0671 0.0568 0.0523
#> Fold 3 -0.0631 0.0616
#> Fold 4 0.0659 -0.0549 0.0527 0.0527
#> Fold 5 0.0425 0.0672 0.0613 0.0493
#> Fold 6 0.0559 -0.0629 0.0498 0.0487
#> Fold 7 0.0611 0.0472
#> Fold 8 -0.0719 0.0586
#> Fold 9 0.0525
#> Fold 10 -0.0580 0.0603
#> X.25 X.4 X.64 X.81 X.97 X.11 X.2 X.33 X.47
#> Fold 1 0.0604 0.0575
#> Fold 2 0.0478 0.0532 0.0518
#> Fold 3 0.0574 0.0473
#> Fold 4 0.0628
#> Fold 5 0.0518
#> Fold 6 0.0521
#> Fold 7 0.0550
#> Fold 8
#> Fold 9 0.0556 0.0447
#> Fold 10 0.0516
#> X.6 X.72 X.73 X.77 X.79 X.88
#> Fold 1 0.0476
#> Fold 2 0.0514
#> Fold 3
#> Fold 4 -0.0473
#> Fold 5 0.0586 0.07
#> Fold 6 -0.0489
#> Fold 7
#> Fold 8
#> Fold 9
#> Fold 10Polynomial regression for the Auto data revisited: meta cross-validation
In the introductory vignette on cross-validating regression models, following James, Witten, Hastie, & Tibshirani (2021, secs. 5.1, 5.3), we fit polynomial regressions up to degree 10 to the relationship ofmpg to
horsepower for the Auto data, saving the
results in m.1 through m.10. We then used
cv() to compare the cross-validated MSE for the 10 models,
discovering that the 7th degree polynomial had the smallest MSE (by a
small margin); repeating the relevant graph:
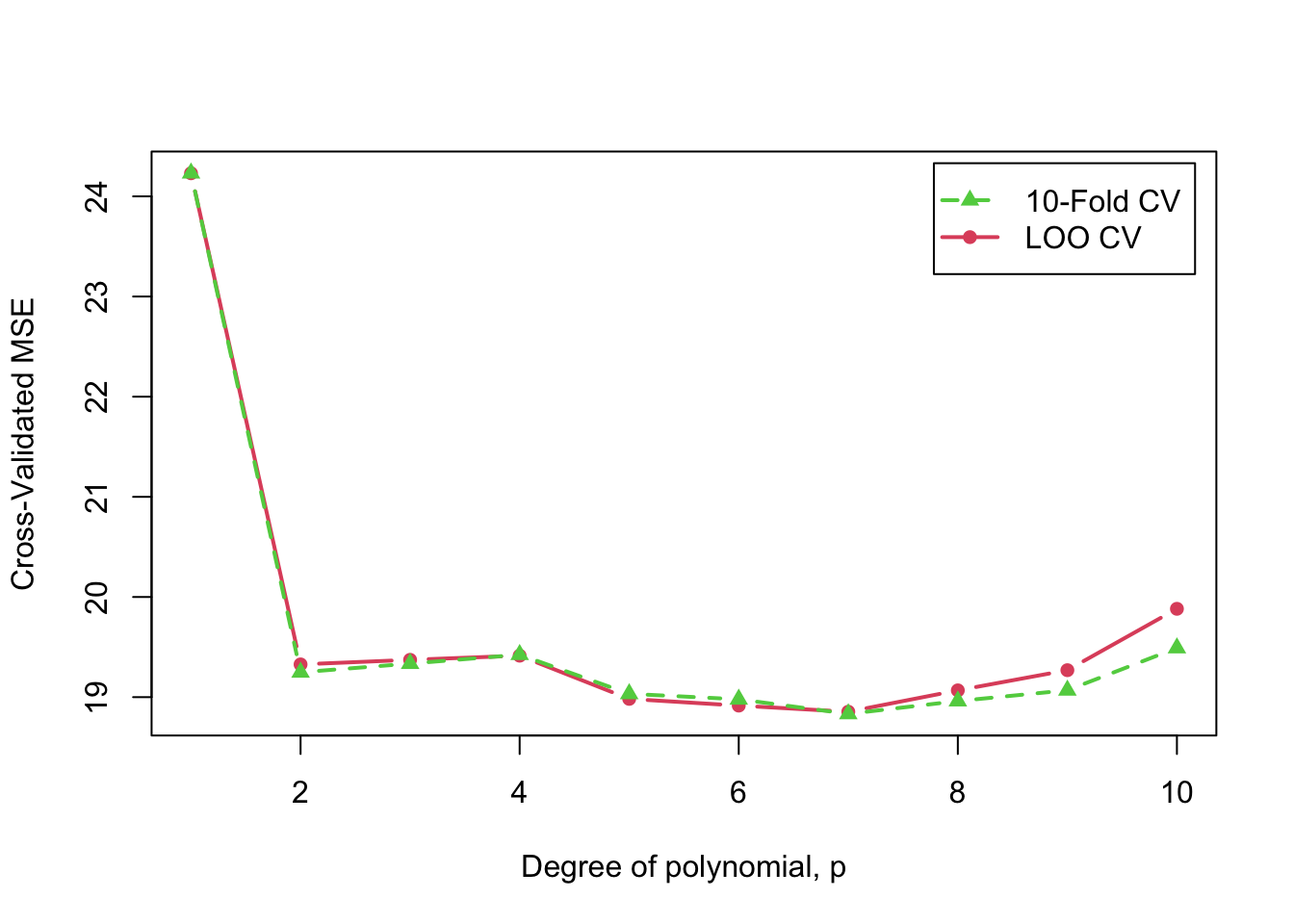
Cross-validated 10-fold and LOO MSE as a function of polynomial degree, (repeated)
If we then select the 7th degree polynomial model, intending to use
it for prediction, the CV estimate of the MSE for this model will be
optimistic. One solution is to cross-validate the process of using CV to
select the “best” model—that is, to apply CV to CV recursively, a
process that we term “meta cross-validation.” The function
selectModelList(), which is suitable for use with
cv(), implements this idea.
Applying selectModelList() to the Auto
polynomial-regression models, and using 10-fold CV, we obtain:
metaCV.auto <- cv(
selectModelList,
Auto,
working.model = models(m.1, m.2, m.3, m.4, m.5,
m.6, m.7, m.8, m.9, m.10),
save.model = TRUE,
seed = 2120
)
#> R RNG seed set to 2120
summary(metaCV.auto)
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 19.105
#> bias-adjusted cross-validation criterion = 19.68
#> full-sample criterion = 18.746
(m.sel <- cvInfo(metaCV.auto, "selected model"))
#>
#> Call:
#> lm(formula = mpg ~ poly(horsepower, 7), data = Auto)
#>
#> Coefficients:
#> (Intercept) poly(horsepower, 7)1 poly(horsepower, 7)2
#> 23.45 -120.14 44.09
#> poly(horsepower, 7)3 poly(horsepower, 7)4 poly(horsepower, 7)5
#> -3.95 -5.19 13.27
#> poly(horsepower, 7)6 poly(horsepower, 7)7
#> -8.55 7.98
cv(m.sel, seed = 2120) # same seed for same folds
#> R RNG seed set to 2120
#> cross-validation criterion (mse) = 18.898As expected, meta CV produces a larger estimate of MSE for the selected 7th degree polynomial model than CV applied directly to this model.
We can equivalently call cv() with the list of models as
the first argument and set meta=TRUE:
Mroz’s logistic regression revisited
Next, let’s apply model selection to Mroz’s logistic regression for
married women’s labor-force participation, also discussed in the
introductory vignette on cross-validating regression models. First,
recall the logistic regression model that we fit to the
Mroz data:
data("Mroz", package = "carData")
m.mroz <- glm(lfp ~ ., data = Mroz, family = binomial)
summary(m.mroz)
#>
#> Call:
#> glm(formula = lfp ~ ., family = binomial, data = Mroz)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 3.18214 0.64438 4.94 7.9e-07 ***
#> k5 -1.46291 0.19700 -7.43 1.1e-13 ***
#> k618 -0.06457 0.06800 -0.95 0.34234
#> age -0.06287 0.01278 -4.92 8.7e-07 ***
#> wcyes 0.80727 0.22998 3.51 0.00045 ***
#> hcyes 0.11173 0.20604 0.54 0.58762
#> lwg 0.60469 0.15082 4.01 6.1e-05 ***
#> inc -0.03445 0.00821 -4.20 2.7e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 1029.75 on 752 degrees of freedom
#> Residual deviance: 905.27 on 745 degrees of freedom
#> AIC: 921.3
#>
#> Number of Fisher Scoring iterations: 4Applying stepwise model selection Mroz’s logistic regression, using
BIC as the model-selection criterion (via the argument
k=log(nrow(Mroz)) to stepAIC()) selects 5 of
the 7 original predictors:
m.mroz.sel <- stepAIC(m.mroz, k = log(nrow(Mroz)),
trace = FALSE)
summary(m.mroz.sel)
#>
#> Call:
#> glm(formula = lfp ~ k5 + age + wc + lwg + inc, family = binomial,
#> data = Mroz)
#>
#> Coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 2.9019 0.5429 5.35 9.0e-08 ***
#> k5 -1.4318 0.1932 -7.41 1.3e-13 ***
#> age -0.0585 0.0114 -5.13 2.9e-07 ***
#> wcyes 0.8724 0.2064 4.23 2.4e-05 ***
#> lwg 0.6157 0.1501 4.10 4.1e-05 ***
#> inc -0.0337 0.0078 -4.32 1.6e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for binomial family taken to be 1)
#>
#> Null deviance: 1029.75 on 752 degrees of freedom
#> Residual deviance: 906.46 on 747 degrees of freedom
#> AIC: 918.5
#>
#> Number of Fisher Scoring iterations: 3
BayesRule(Mroz$lfp == "yes",
predict(m.mroz.sel, type = "response"))
#> [1] 0.31873
#> attr(,"casewise loss")
#> [1] "y != round(yhat)"Bayes rule applied to the selected model misclassifies 32% of the
cases in the Mroz data.
Cross-validating the selected model produces a similar, slightly larger, estimate of misclassification, about 33%:
summary(cv(m.mroz.sel, criterion = BayesRule, seed = 345266))
#> R RNG seed set to 345266
#> 10-Fold Cross Validation
#> method: exact
#> criterion: BayesRule
#> cross-validation criterion = 0.33068
#> bias-adjusted cross-validation criterion = 0.33332
#> 95% CI for bias-adjusted CV criterion = (0.2997, 0.36695)
#> full-sample criterion = 0.31873Is this estimate of predictive performance optimistic?
We proceed to apply the model-selection procedure by cross-validation, producing more or less the same result:
m.mroz.sel.cv <- cv(
selectStepAIC,
Mroz,
seed = 6681,
criterion = BayesRule,
working.model = m.mroz,
AIC = FALSE
)
#> R RNG seed set to 6681
summary(m.mroz.sel.cv)
#> 10-Fold Cross Validation
#> criterion: BayesRule
#> cross-validation criterion = 0.33068
#> bias-adjusted cross-validation criterion = 0.33452
#> 95% CI for bias-adjusted CV criterion = (0.3009, 0.36815)
#> full-sample criterion = 0.31873Setting AIC=FALSE in the call to cv() uses
the BIC rather than the AIC as the model-selection criterion. As it
turns out, exactly the same predictors are selected when each of the 10
folds are omitted, and the several coefficient estimates are very
similar, as we show using compareFolds():
compareFolds(m.mroz.sel.cv)
#> CV criterion by folds:
#> fold.1 fold.2 fold.3 fold.4 fold.5 fold.6 fold.7 fold.8 fold.9 fold.10
#> 0.27632 0.40789 0.34211 0.36000 0.28000 0.37333 0.32000 0.29333 0.33333 0.32000
#>
#> Coefficients by folds:
#> (Intercept) age inc k5 lwg wcyes
#> Fold 1 2.5014 -0.0454 -0.0388 -1.3613 0.5653 0.85
#> Fold 2 3.0789 -0.0659 -0.0306 -1.5335 0.6923 0.79
#> Fold 3 3.0141 -0.0595 -0.0305 -1.3994 0.5428 0.86
#> Fold 4 2.7251 -0.0543 -0.0354 -1.4474 0.6298 1.09
#> Fold 5 2.7617 -0.0566 -0.0320 -1.4752 0.6324 0.74
#> Fold 6 3.0234 -0.0621 -0.0348 -1.4537 0.6618 0.94
#> Fold 7 2.9615 -0.0600 -0.0351 -1.4127 0.5835 0.97
#> Fold 8 2.9598 -0.0603 -0.0329 -1.3865 0.6210 0.69
#> Fold 9 3.2481 -0.0650 -0.0381 -1.4138 0.6093 0.94
#> Fold 10 2.7724 -0.0569 -0.0295 -1.4503 0.6347 0.85In this example, therefore, we appear to obtain a realistic estimate of model performance directly from the selected model, because there is little added uncertainty induced by model selection.
Cross-validating choice of transformations in regression
The cv package also provides a cv()
procedure, selectTrans(), for choosing transformations of
the predictors and the response in regression.
Some background: As Weisberg (2014, sec. 8.2) explains, there are technical advantages to having (numeric) predictors in linear regression analysis that are themselves linearly related. If the predictors aren’t linearly related, then the relationships between them can often be straightened by power transformations. Transformations can be selected after graphical examination of the data, or by analytic methods. Once the relationships between the predictors are linearized, it can be advantageous similarly to transform the response variable towards normality.
Selecting transformations analytically raises the possibility of automating the process, as would be required for cross-validation. One could, in principle, apply graphical methods to select transformations for each fold, but because a data analyst couldn’t forget the choices made for previous folds, the process wouldn’t really be applied independently to the folds.
To illustrate, we adapt an example appearing in several places in
Fox & Weisberg (2019) (for example in
Chapter 3 on transforming data), using data on the prestige and other
characteristics of 102 Canadian occupations circa 1970. The data are in
the Prestige data frame in the carData
package:
data("Prestige", package = "carData")
head(Prestige)
#> education income women prestige census type
#> gov.administrators 13.11 12351 11.16 68.8 1113 prof
#> general.managers 12.26 25879 4.02 69.1 1130 prof
#> accountants 12.77 9271 15.70 63.4 1171 prof
#> purchasing.officers 11.42 8865 9.11 56.8 1175 prof
#> chemists 14.62 8403 11.68 73.5 2111 prof
#> physicists 15.64 11030 5.13 77.6 2113 prof
summary(Prestige)
#> education income women prestige census
#> Min. : 6.38 Min. : 611 Min. : 0.00 Min. :14.8 Min. :1113
#> 1st Qu.: 8.45 1st Qu.: 4106 1st Qu.: 3.59 1st Qu.:35.2 1st Qu.:3120
#> Median :10.54 Median : 5930 Median :13.60 Median :43.6 Median :5135
#> Mean :10.74 Mean : 6798 Mean :28.98 Mean :46.8 Mean :5402
#> 3rd Qu.:12.65 3rd Qu.: 8187 3rd Qu.:52.20 3rd Qu.:59.3 3rd Qu.:8312
#> Max. :15.97 Max. :25879 Max. :97.51 Max. :87.2 Max. :9517
#> type
#> bc :44
#> prof:31
#> wc :23
#> NA's: 4
#>
#> The variables in the Prestige data set are:
-
education: average years of education for incumbents in the occupation, from the 1971 Canadian Census. -
income: average dollars of annual income for the occupation, from the Census. -
women: percentage of occupational incumbents who were women, also from the Census. -
prestige: the average prestige rating of the occupation on a 0–100 “thermometer” scale, in a Canadian social survey conducted around the same time. -
type, type of occupation, andcensus, the Census occupational code, which are not used in our example.
The object of a regression analysis for the Prestige
data (and their original purpose) is to predict occupational prestige
from the other variables in the data set.
A scatterplot matrix (using the scatterplotMatrix()
function in the car package) of the numeric variables
in the data reveals that the distributions of income and
women are positively skewed, and that some of the
relationships among the three predictors, and between the predictors and
the response (i.e., prestige), are nonlinear:
library("car")
#> Loading required package: carData
scatterplotMatrix(
~ prestige + income + education + women,
data = Prestige,
smooth = list(spread = FALSE)
)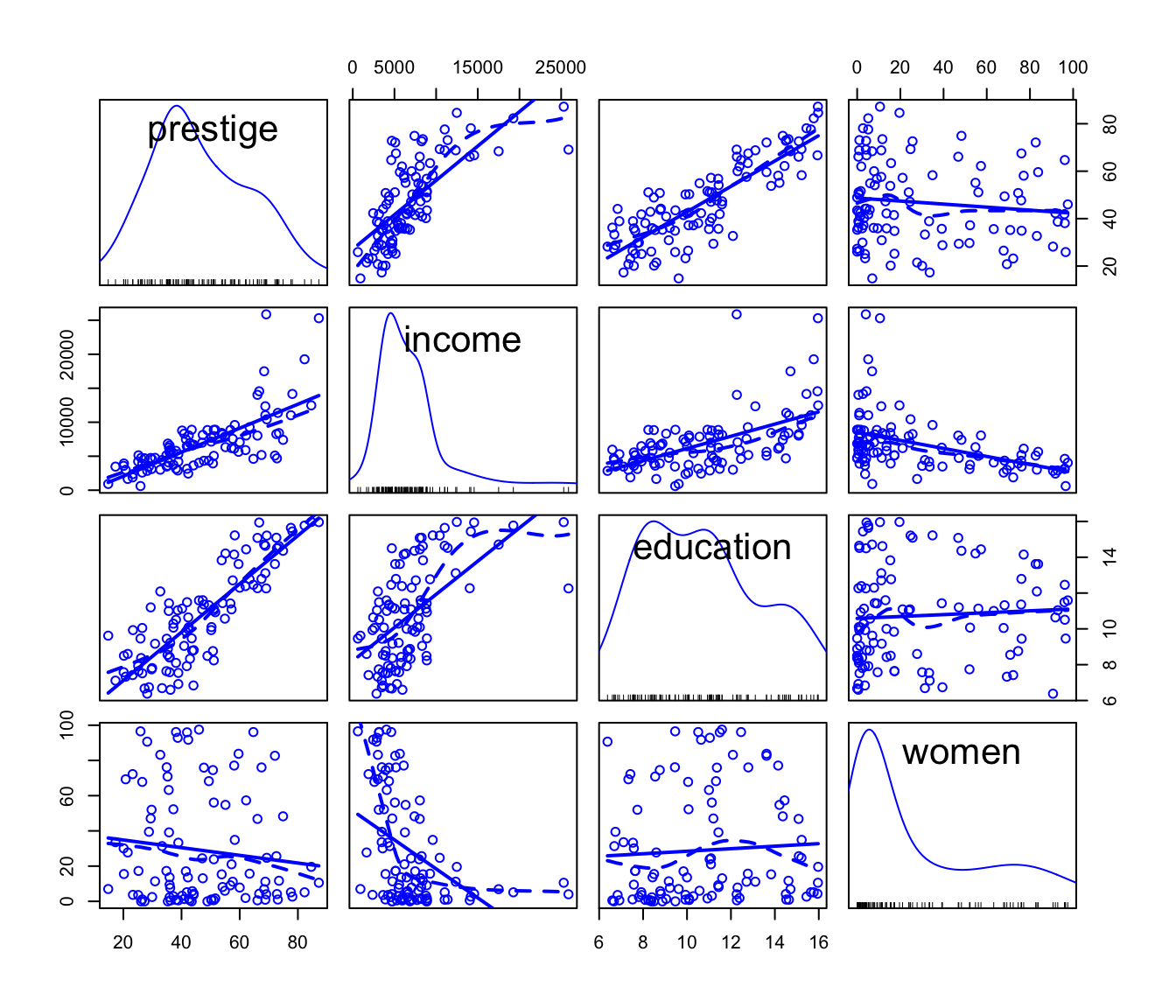
Scatterplot matrix for the Prestige data.
The powerTransform() function in the
car package transforms variables towards multivariate
normality by a generalization of Box and Cox’s maximum-likelihood-like
approach (Box & Cox, 1964). Several
“families” of power transformations can be used, including the original
Box-Cox family, simple powers (and roots), and two adaptations of the
Box-Cox family to data that may include negative values and zeros: the
Box-Cox-with-negatives family and the Yeo-Johnson family; see Weisberg (2014, Chapter 8), and Fox & Weisberg (2019, Chapter 3) for
details. Because women has some zero values, we use the
Yeo-Johnson family:
trans <- powerTransform(cbind(income, education, women) ~ 1,
data = Prestige,
family = "yjPower")
summary(trans)
#> yjPower Transformations to Multinormality
#> Est Power Rounded Pwr Wald Lwr Bnd Wald Upr Bnd
#> income 0.2678 0.33 0.1051 0.4304
#> education 0.5162 1.00 -0.2822 1.3145
#> women 0.1630 0.16 0.0112 0.3149
#>
#> Likelihood ratio test that all transformation parameters are equal to 0
#> LRT df pval
#> LR test, lambda = (0 0 0) 15.739 3 0.00128We thus have evidence of the desirability of transforming
income (by the
power) and women (by the
power—which is close to the “0” power, i.e., the log transformation),
but not education. Applying the “rounded” power
transformations makes the predictors better-behaved:
P <- Prestige[, c("prestige", "income", "education", "women")]
(lambdas <- trans$roundlam)
#> income education women
#> 0.33000 1.00000 0.16302
names(lambdas) <- c("income", "education", "women")
for (var in c("income", "education", "women")) {
P[, var] <- yjPower(P[, var], lambda = lambdas[var])
}
summary(P)
#> prestige income education women
#> Min. :14.8 Min. :22.2 Min. : 6.38 Min. :0.00
#> 1st Qu.:35.2 1st Qu.:44.2 1st Qu.: 8.45 1st Qu.:1.73
#> Median :43.6 Median :50.3 Median :10.54 Median :3.36
#> Mean :46.8 Mean :50.8 Mean :10.74 Mean :3.50
#> 3rd Qu.:59.3 3rd Qu.:56.2 3rd Qu.:12.65 3rd Qu.:5.59
#> Max. :87.2 Max. :83.6 Max. :15.97 Max. :6.83
scatterplotMatrix(
~ prestige + income + education + women,
data = P,
smooth = list(spread = FALSE)
)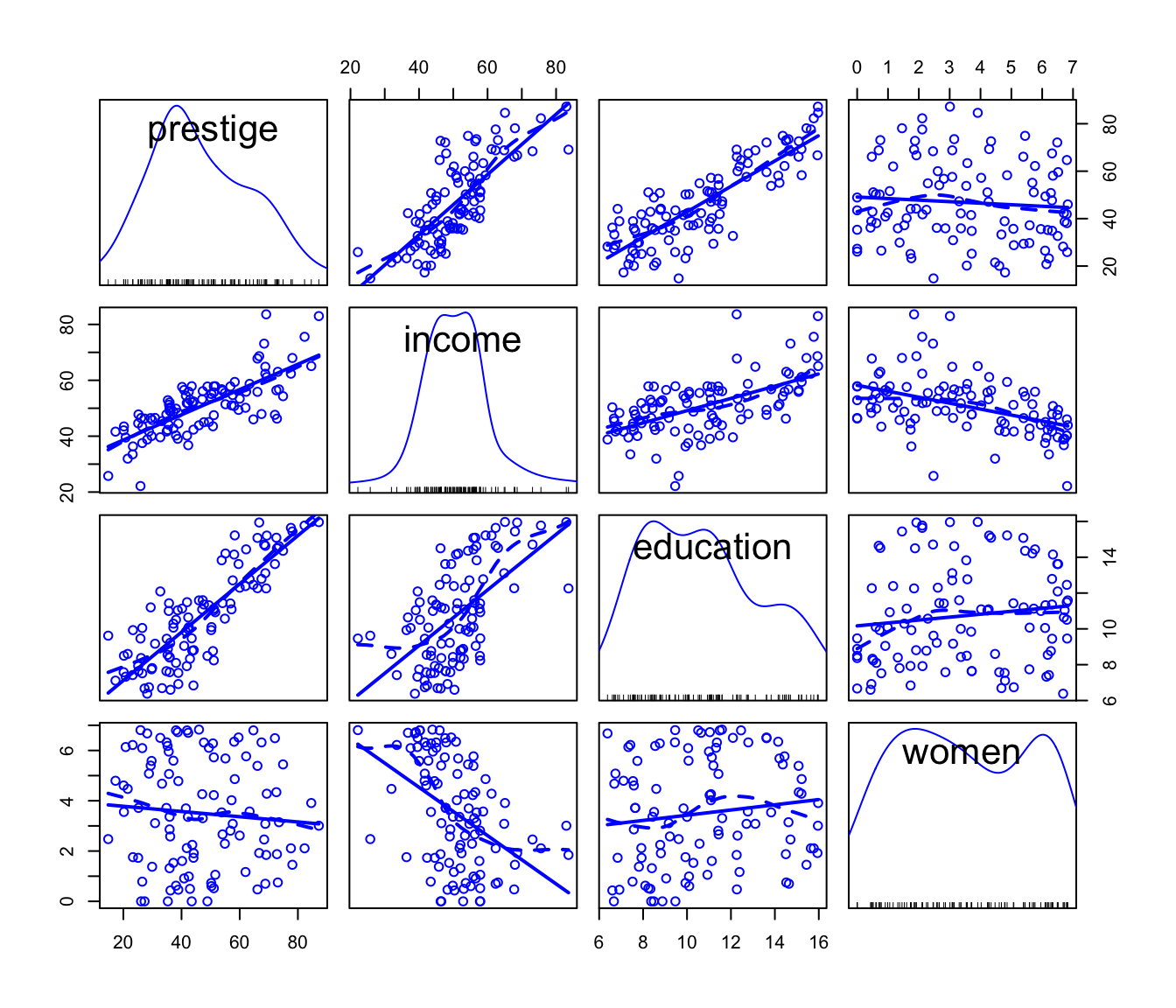
Scatterplot matrix for the Prestige data with the
predictors transformed.
Comparing the MSE for the regressions with the original and transformed predictors shows a advantage to the latter:
m.pres <- lm(prestige ~ income + education + women, data = Prestige)
m.pres.trans <- lm(prestige ~ income + education + women, data = P)
mse(Prestige$prestige, fitted(m.pres))
#> [1] 59.153
#> attr(,"casewise loss")
#> [1] "(y - yhat)^2"
mse(P$prestige, fitted(m.pres.trans))
#> [1] 50.6
#> attr(,"casewise loss")
#> [1] "(y - yhat)^2"Similarly, component+residual plots for the two regressions, produced
by the crPlots() function in the car
package, suggest that the partial relationship of prestige
to income is more nearly linear in the transformed data,
but the transformation of women fails to capture what
appears to be a slight quadratic partial relationship; the partial
relationship of prestige to education is close
to linear in both regressions:
crPlots(m.pres)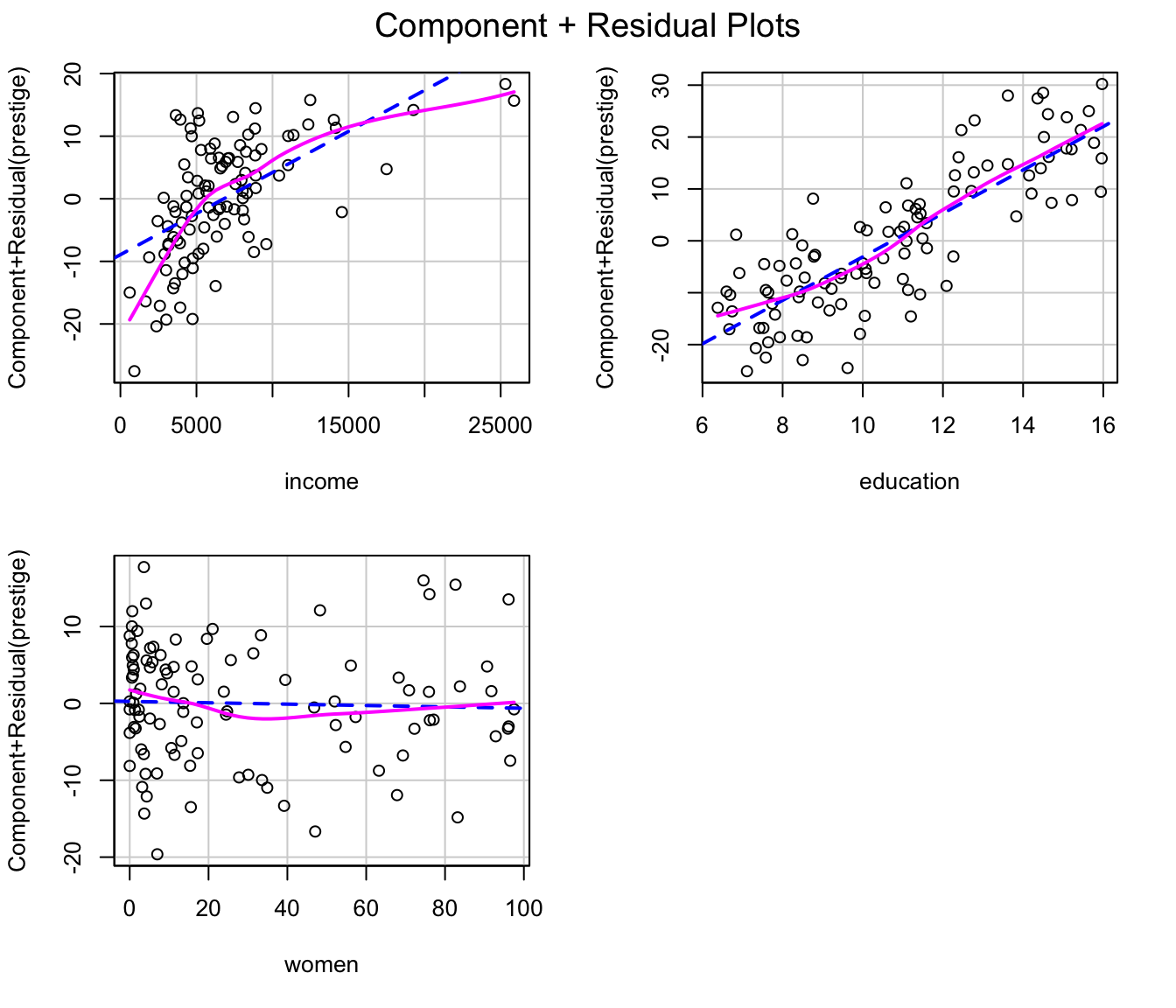
Component+residual plots for the Prestige regression with
the original predictors.
crPlots(m.pres.trans)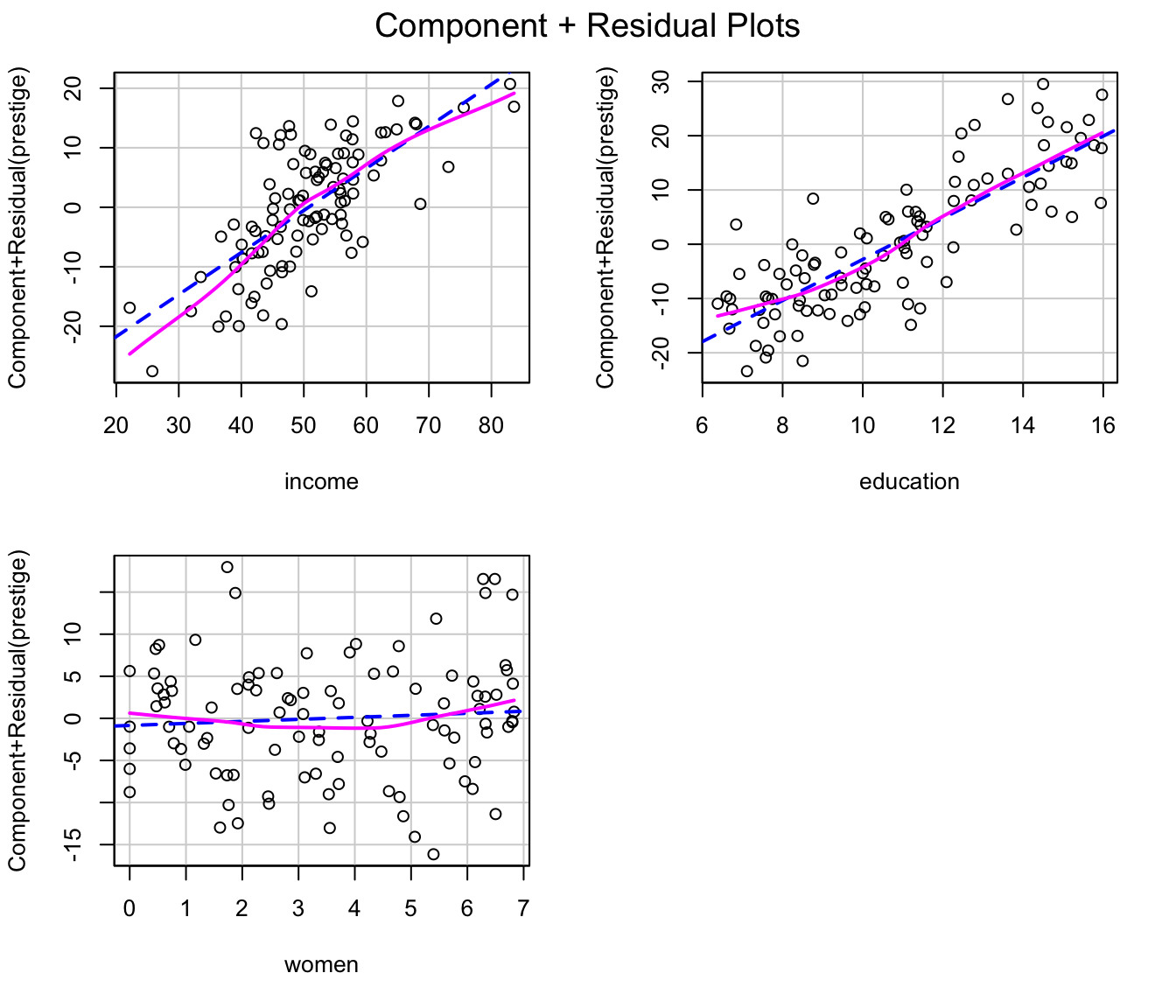
Component+residual plots for the Prestige regression with
transformed predictors.
Having transformed the predictors towards multinormality, we now
consider whether there’s evidence for transforming the response (using
powerTransform() for Box and Cox’s original method), and we
discover that there’s not:
summary(powerTransform(m.pres.trans))
#> bcPower Transformation to Normality
#> Est Power Rounded Pwr Wald Lwr Bnd Wald Upr Bnd
#> Y1 1.0194 1 0.6773 1.3615
#>
#> Likelihood ratio test that transformation parameter is equal to 0
#> (log transformation)
#> LRT df pval
#> LR test, lambda = (0) 32.217 1 1.38e-08
#>
#> Likelihood ratio test that no transformation is needed
#> LRT df pval
#> LR test, lambda = (1) 0.012384 1 0.911The selectTrans() function in the cv
package automates the process of selecting predictor and response
transformations. The function takes a data set and
“working” model as arguments, along with the candidate
predictors and response for transformation,
and the transformation family to employ. If the
predictors argument is missing then only the response is
transformed, and if the response argument is missing, only
the supplied predictors are transformed. The default family
for transforming the predictors is "bcPower"—the original
Box-Cox family—as is the default family.y for transforming
the response; here we specify family="yjPower because of
the zeros in women. selectTrans() returns the
result of applying a lack-of-fit criterion to the model after the
selected transformation is applied, with the default
criterion=mse:
selectTrans(
data = Prestige,
model = m.pres,
predictors = c("income", "education", "women"),
response = "prestige",
family = "yjPower"
)
#> $criterion
#> [1] 50.6
#> attr(,"casewise loss")
#> [1] "(y - yhat)^2"
#>
#> $model
#> NULLselectTrans() also takes an optional
indices argument, making it suitable for doing computations
on a subset of the data (i.e., a CV fold), and hence for use with
cv() (see ?selectTrans for details):
cvs <- cv(
selectTrans,
data = Prestige,
working.model = m.pres,
seed = 1463,
predictors = c("income", "education", "women"),
response = "prestige",
family = "yjPower"
)
#> R RNG seed set to 1463
summary(cvs)
#> 10-Fold Cross Validation
#> criterion: mse
#> cross-validation criterion = 54.487
#> bias-adjusted cross-validation criterion = 54.308
#> full-sample criterion = 50.6
cv(m.pres, seed = 1463) # untransformed model with same folds
#> R RNG seed set to 1463
#> cross-validation criterion (mse) = 63.293
compareFolds(cvs)
#> CV criterion by folds:
#> fold.1 fold.2 fold.3 fold.4 fold.5 fold.6 fold.7 fold.8 fold.9 fold.10
#> 63.453 79.257 20.634 94.569 19.902 55.821 26.555 75.389 55.702 50.215
#>
#> Coefficients by folds:
#> lam.education lam.income lam.women lambda
#> Fold 1 1.000 0.330 0.330 1
#> Fold 2 1.000 0.330 0.169 1
#> Fold 3 1.000 0.330 0.330 1
#> Fold 4 1.000 0.330 0.330 1
#> Fold 5 1.000 0.330 0.000 1
#> Fold 6 1.000 0.330 0.330 1
#> Fold 7 1.000 0.330 0.330 1
#> Fold 8 1.000 0.330 0.000 1
#> Fold 9 1.000 0.330 0.000 1
#> Fold 10 1.000 0.330 0.000 1The results suggest that the predictive power of the transformed
regression is reliably greater than that of the untransformed regression
(though in both case, the cross-validated MSE is considerably higher
than the MSE computed for the whole data). Examining the selected
transformations for each fold reveals that the predictor
education and the response prestige are never
transformed; that the
power is selected for income in all of the folds; and that
the transformation selected for women varies narrowly
across the folds between the
th
power (i.e., log) and the
power.
Selecting both transformations and predictors2
As we mentioned, Hastie et al. (2009, sec. 7.10.2: “The Wrong and Right Way to Do Cross-validation”) explain that honest cross-validation has to take account of model specification and selection. Statistical modeling is at least partly a craft, and one could imagine applying that craft to successive partial data sets, each with a fold removed. The resulting procedure would be tedious, though possibly worth the effort, but it would also be difficult to realize in practice: After all, we can hardly erase our memory of statistical modeling choices between analyzing partial data sets.
Alternatively, if we’re able to automate the process of model
selection, then we can more realistically apply CV mechanically. That’s
what we did in the preceding two sections, first for predictor selection
and then for selection of transformations in regression. In this
section, we consider the case where we both select variable
transformations and then proceed to select predictors. It’s insufficient
to apply these steps sequentially, first, for example, using
cv() with selectTrans() and then with
selectStepAIC(); rather we should apply the whole
model-selection procedure with each fold omitted. The
selectTransAndStepAIC() function, also supplied by the
cv package, does exactly that.
To illustrate this process, we return to the Auto data
set:
summary(Auto)
#> mpg cylinders displacement horsepower weight
#> Min. : 9.0 Min. :3.00 Min. : 68 Min. : 46.0 Min. :1613
#> 1st Qu.:17.0 1st Qu.:4.00 1st Qu.:105 1st Qu.: 75.0 1st Qu.:2225
#> Median :22.8 Median :4.00 Median :151 Median : 93.5 Median :2804
#> Mean :23.4 Mean :5.47 Mean :194 Mean :104.5 Mean :2978
#> 3rd Qu.:29.0 3rd Qu.:8.00 3rd Qu.:276 3rd Qu.:126.0 3rd Qu.:3615
#> Max. :46.6 Max. :8.00 Max. :455 Max. :230.0 Max. :5140
#>
#> acceleration year origin name
#> Min. : 8.0 Min. :70 Min. :1.00 amc matador : 5
#> 1st Qu.:13.8 1st Qu.:73 1st Qu.:1.00 ford pinto : 5
#> Median :15.5 Median :76 Median :1.00 toyota corolla : 5
#> Mean :15.5 Mean :76 Mean :1.58 amc gremlin : 4
#> 3rd Qu.:17.0 3rd Qu.:79 3rd Qu.:2.00 amc hornet : 4
#> Max. :24.8 Max. :82 Max. :3.00 chevrolet chevette: 4
#> (Other) :365
xtabs( ~ year, data = Auto)
#> year
#> 70 71 72 73 74 75 76 77 78 79 80 81 82
#> 29 27 28 40 26 30 34 28 36 29 27 28 30
xtabs( ~ origin, data = Auto)
#> origin
#> 1 2 3
#> 245 68 79
xtabs( ~ cylinders, data = Auto)
#> cylinders
#> 3 4 5 6 8
#> 4 199 3 83 103We previously used the Auto here in a preliminary
example where we employed CV to inform the selection of the order of a
polynomial regression of mpg on horsepower.
Here, we consider more generally the problem of predicting
mpg from the other variables in the Auto data.
We begin with a bit of data management, and then examine the pairwise
relationships among the numeric variables in the data set:
Auto$cylinders <- factor(Auto$cylinders,
labels = c("3.4", "3.4", "5.6", "5.6", "8"))
Auto$year <- as.factor(Auto$year)
Auto$origin <- factor(Auto$origin,
labels = c("America", "Europe", "Japan"))
rownames(Auto) <- make.names(Auto$name, unique = TRUE)
Auto$name <- NULL
scatterplotMatrix(
~ mpg + displacement + horsepower + weight + acceleration,
smooth = list(spread = FALSE),
data = Auto
)
Scatterplot matrix for the numeric variables in the Auto
data
A comment before we proceed: origin is clearly
categorical and so converting it to a factor is natural, but we could
imagine treating cylinders and year as numeric
predictors. There are, however, only 5 distinct values of
cylinders (ranging from 3 to 8), but cars with 3 or 5
cylinders are rare. and none of the cars has 7 cylinders. There are
similarly only 13 distinct years between 1970 and 1982 in the data, and
the relationship between mpg and year is
difficult to characterize.3 It’s apparent that most these variables are
positively skewed and that many of the pairwise relationships among them
are nonlinear.
We begin with a “working model” that specifies linear partial relationships of the response to the numeric predictors:
m.auto <- lm(mpg ~ ., data = Auto)
summary(m.auto)
#>
#> Call:
#> lm(formula = mpg ~ ., data = Auto)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -9.006 -1.745 -0.092 1.525 10.950
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 37.034132 1.969393 18.80 < 2e-16 ***
#> cylinders5.6 -2.602941 0.655200 -3.97 8.5e-05 ***
#> cylinders8 -0.582458 1.171452 -0.50 0.61934
#> displacement 0.017425 0.006734 2.59 0.01004 *
#> horsepower -0.041353 0.013379 -3.09 0.00215 **
#> weight -0.005548 0.000632 -8.77 < 2e-16 ***
#> acceleration 0.061527 0.088313 0.70 0.48643
#> year71 0.968058 0.837390 1.16 0.24841
#> year72 -0.601435 0.825115 -0.73 0.46652
#> year73 -0.687689 0.740272 -0.93 0.35351
#> year74 1.375576 0.876500 1.57 0.11741
#> year75 0.929929 0.859072 1.08 0.27974
#> year76 1.559893 0.822505 1.90 0.05867 .
#> year77 2.909416 0.841729 3.46 0.00061 ***
#> year78 3.175198 0.798940 3.97 8.5e-05 ***
#> year79 5.019299 0.845759 5.93 6.8e-09 ***
#> year80 9.099763 0.897293 10.14 < 2e-16 ***
#> year81 6.688660 0.885218 7.56 3.3e-13 ***
#> year82 8.071125 0.870668 9.27 < 2e-16 ***
#> originEurope 2.046664 0.517124 3.96 9.1e-05 ***
#> originJapan 2.144887 0.507717 4.22 3.0e-05 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 2.92 on 371 degrees of freedom
#> Multiple R-squared: 0.867, Adjusted R-squared: 0.86
#> F-statistic: 121 on 20 and 371 DF, p-value: <2e-16
Anova(m.auto)
#> Anova Table (Type II tests)
#>
#> Response: mpg
#> Sum Sq Df F value Pr(>F)
#> cylinders 292 2 17.09 7.9e-08 ***
#> displacement 57 1 6.70 0.0100 *
#> horsepower 82 1 9.55 0.0021 **
#> weight 658 1 76.98 < 2e-16 ***
#> acceleration 4 1 0.49 0.4864
#> year 3017 12 29.40 < 2e-16 ***
#> origin 190 2 11.13 2.0e-05 ***
#> Residuals 3173 371
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
crPlots(m.auto)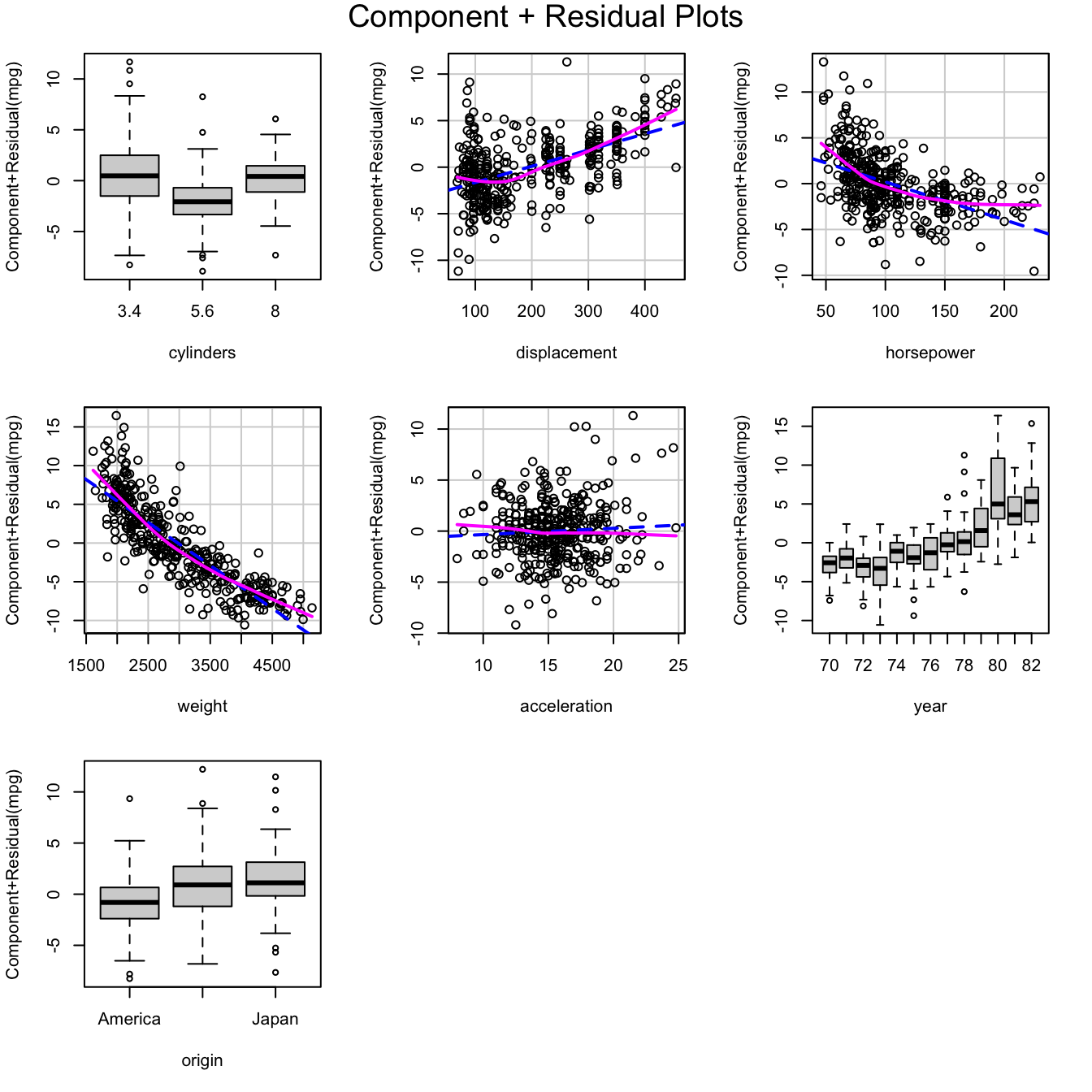
Component+residual plots for the working model fit to the
Auto data
The component+residual plots, created with the crPlots()
function in the previously loaded car package, clearly
reveal the inadequacy of the model.
We proceed to transform the numeric predictors towards multi-normality:
num.predictors <-
c("displacement", "horsepower", "weight", "acceleration")
tr.x <- powerTransform(Auto[, num.predictors])
summary(tr.x)
#> bcPower Transformations to Multinormality
#> Est Power Rounded Pwr Wald Lwr Bnd Wald Upr Bnd
#> displacement -0.0509 0 -0.2082 0.1065
#> horsepower -0.1249 0 -0.2693 0.0194
#> weight -0.0870 0 -0.2948 0.1208
#> acceleration 0.3061 0 -0.0255 0.6376
#>
#> Likelihood ratio test that transformation parameters are equal to 0
#> (all log transformations)
#> LRT df pval
#> LR test, lambda = (0 0 0 0) 4.8729 4 0.301
#>
#> Likelihood ratio test that no transformations are needed
#> LRT df pval
#> LR test, lambda = (1 1 1 1) 390.08 4 <2e-16We then apply the (rounded) transformations—all, as it turns out, logs—to the data and re-estimate the model:
A <- Auto
powers <- tr.x$roundlam
for (pred in num.predictors) {
A[, pred] <- bcPower(A[, pred], lambda = powers[pred])
}
head(A)
#> mpg cylinders displacement horsepower weight
#> chevrolet.chevelle.malibu 18 8 5.7268 4.8675 8.1617
#> buick.skylark.320 15 8 5.8579 5.1059 8.2142
#> plymouth.satellite 18 8 5.7621 5.0106 8.1421
#> amc.rebel.sst 16 8 5.7170 5.0106 8.1412
#> ford.torino 17 8 5.7104 4.9416 8.1458
#> ford.galaxie.500 15 8 6.0615 5.2883 8.3759
#> acceleration year origin
#> chevrolet.chevelle.malibu 2.4849 70 America
#> buick.skylark.320 2.4423 70 America
#> plymouth.satellite 2.3979 70 America
#> amc.rebel.sst 2.4849 70 America
#> ford.torino 2.3514 70 America
#> ford.galaxie.500 2.3026 70 America
m <- update(m.auto, data = A)Finally, we perform Box-Cox regression to transform the response (also obtaining a log transformation):
summary(powerTransform(m))
#> bcPower Transformation to Normality
#> Est Power Rounded Pwr Wald Lwr Bnd Wald Upr Bnd
#> Y1 0.0024 0 -0.1607 0.1654
#>
#> Likelihood ratio test that transformation parameter is equal to 0
#> (log transformation)
#> LRT df pval
#> LR test, lambda = (0) 0.00080154 1 0.977
#>
#> Likelihood ratio test that no transformation is needed
#> LRT df pval
#> LR test, lambda = (1) 124.13 1 <2e-16
m <- update(m, log(mpg) ~ .)
summary(m)
#>
#> Call:
#> lm(formula = log(mpg) ~ cylinders + displacement + horsepower +
#> weight + acceleration + year + origin, data = A)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.3341 -0.0577 0.0041 0.0607 0.3808
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 8.8965 0.3582 24.84 < 2e-16 ***
#> cylinders5.6 -0.0636 0.0257 -2.47 0.014 *
#> cylinders8 -0.0769 0.0390 -1.97 0.049 *
#> displacement 0.0280 0.0515 0.54 0.587
#> horsepower -0.2901 0.0563 -5.15 4.2e-07 ***
#> weight -0.5427 0.0819 -6.62 1.2e-10 ***
#> acceleration -0.1421 0.0563 -2.52 0.012 *
#> year71 0.0250 0.0289 0.87 0.387
#> year72 -0.0168 0.0289 -0.58 0.562
#> year73 -0.0426 0.0260 -1.64 0.103
#> year74 0.0493 0.0304 1.62 0.106
#> year75 0.0472 0.0296 1.59 0.112
#> year76 0.0709 0.0284 2.49 0.013 *
#> year77 0.1324 0.0293 4.52 8.2e-06 ***
#> year78 0.1447 0.0278 5.21 3.1e-07 ***
#> year79 0.2335 0.0292 7.99 1.7e-14 ***
#> year80 0.3238 0.0317 10.22 < 2e-16 ***
#> year81 0.2565 0.0309 8.29 2.1e-15 ***
#> year82 0.3076 0.0304 10.13 < 2e-16 ***
#> originEurope 0.0492 0.0195 2.52 0.012 *
#> originJapan 0.0441 0.0195 2.26 0.024 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.104 on 371 degrees of freedom
#> Multiple R-squared: 0.911, Adjusted R-squared: 0.906
#> F-statistic: 189 on 20 and 371 DF, p-value: <2e-16
Anova(m)
#> Anova Table (Type II tests)
#>
#> Response: log(mpg)
#> Sum Sq Df F value Pr(>F)
#> cylinders 0.07 2 3.05 0.048 *
#> displacement 0.00 1 0.30 0.587
#> horsepower 0.29 1 26.54 4.2e-07 ***
#> weight 0.48 1 43.88 1.2e-10 ***
#> acceleration 0.07 1 6.37 0.012 *
#> year 4.45 12 34.13 < 2e-16 ***
#> origin 0.08 2 3.71 0.025 *
#> Residuals 4.03 371
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The transformed numeric variables are much better-behaved:
scatterplotMatrix(
~ log(mpg) + displacement + horsepower + weight
+ acceleration,
smooth = list(spread = FALSE),
data = A
)
Scatterplot matrix for the transformed numeric variables in the
Auto data
And the partial relationships in the model fit to the transformed data are much more nearly linear:
crPlots(m)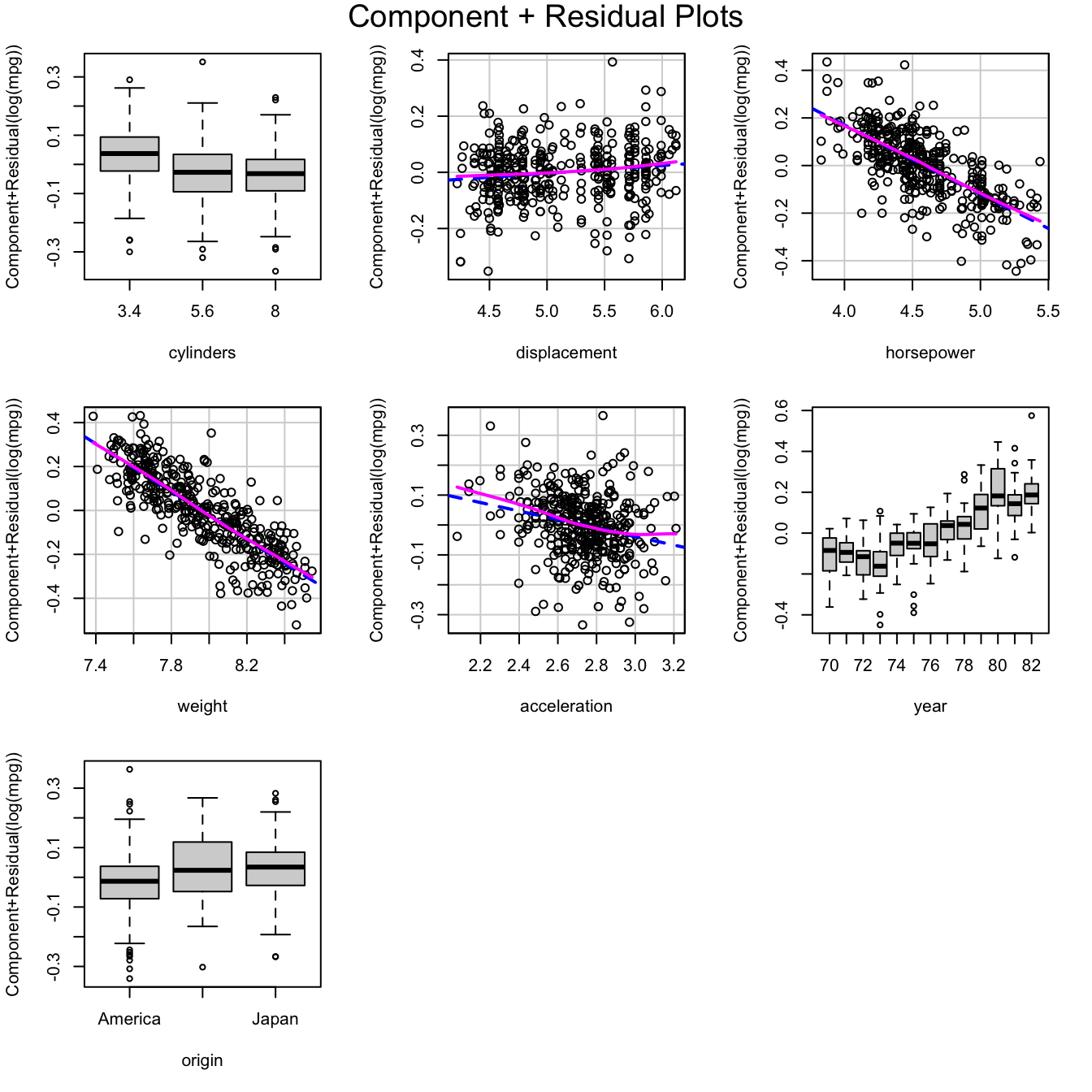
Component+residual plots for the model fit to the transformed
Auto data
Having transformed both the numeric predictors and the response, we
proceed to use the stepAIC() function in the
MASS package to perform predictor selection, employing
the BIC model-selection criterion (by setting the k
argument of stepAIC() to
):
m.step <- stepAIC(m, k=log(nrow(A)), trace=FALSE)
summary(m.step)
#>
#> Call:
#> lm(formula = log(mpg) ~ horsepower + weight + acceleration +
#> year + origin, data = A)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.3523 -0.0568 0.0068 0.0674 0.3586
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 9.43459 0.26153 36.07 < 2e-16 ***
#> horsepower -0.27625 0.05614 -4.92 1.3e-06 ***
#> weight -0.60907 0.05600 -10.88 < 2e-16 ***
#> acceleration -0.13138 0.05319 -2.47 0.01397 *
#> year71 0.02798 0.02894 0.97 0.33412
#> year72 -0.00711 0.02845 -0.25 0.80274
#> year73 -0.03953 0.02601 -1.52 0.12947
#> year74 0.05275 0.02999 1.76 0.07936 .
#> year75 0.05320 0.02928 1.82 0.07004 .
#> year76 0.07432 0.02821 2.63 0.00878 **
#> year77 0.13793 0.02888 4.78 2.6e-06 ***
#> year78 0.14588 0.02753 5.30 2.0e-07 ***
#> year79 0.23604 0.02908 8.12 7.0e-15 ***
#> year80 0.33527 0.03115 10.76 < 2e-16 ***
#> year81 0.26287 0.03056 8.60 < 2e-16 ***
#> year82 0.32339 0.02961 10.92 < 2e-16 ***
#> originEurope 0.05582 0.01678 3.33 0.00097 ***
#> originJapan 0.04355 0.01748 2.49 0.01314 *
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 0.105 on 374 degrees of freedom
#> Multiple R-squared: 0.909, Adjusted R-squared: 0.905
#> F-statistic: 220 on 17 and 374 DF, p-value: <2e-16
Anova(m.step)
#> Anova Table (Type II tests)
#>
#> Response: log(mpg)
#> Sum Sq Df F value Pr(>F)
#> horsepower 0.27 1 24.21 1.3e-06 ***
#> weight 1.30 1 118.28 < 2e-16 ***
#> acceleration 0.07 1 6.10 0.0140 *
#> year 4.76 12 36.05 < 2e-16 ***
#> origin 0.14 2 6.21 0.0022 **
#> Residuals 4.11 374
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The selected model includes three of the numeric predictors,
horsepower, weight, and
acceleration, along with the factors year and
origin. We can calculate the MSE for this model, but we
expect that the result will be optimistic because we used the whole data
to help specify the model
This is considerably smaller than the MSE for the original working model:
A perhaps subtle point is that we compute the MSE for the selected
model on the original mpg response scale rather than the
log scale, so as to make the selected model comparable to the working
model. That’s slightly uncomfortable given the skewed distribution of
mpg. An alternative is to use the median absolute error
instead of the mean-squared error, employing the
medAbsErr() function from the cv
package:
medAbsErr(Auto$mpg, exp(fitted(m.step)))
#> [1] 1.3396
medAbsErr(Auto$mpg, fitted(m.auto))
#> [1] 1.6661Now let’s use cv() with
selectTransAndStepAIC() to automate and cross-validate the
whole model-specification process:
num.predictors
#> [1] "displacement" "horsepower" "weight" "acceleration"
cvs <- cv(
selectTransStepAIC,
data = Auto,
seed = 76692,
working.model = m.auto,
predictors = num.predictors,
response = "mpg",
AIC = FALSE,
criterion = medAbsErr
)
#> R RNG seed set to 76692
summary(cvs)
#> 10-Fold Cross Validation
#> criterion: medAbsErr
#> cross-validation criterion = 1.4951
#> full-sample criterion = 1.3396
compareFolds(cvs)
#> CV criterion by folds:
#> fold.1 fold.2 fold.3 fold.4 fold.5 fold.6 fold.7 fold.8 fold.9 fold.10
#> 1.5639 1.5766 1.3698 1.2381 1.2461 1.5826 1.3426 1.1905 1.1355 1.5854
#>
#> Coefficients by folds:
#> (Intercept) horsepower lam.acceleration lam.displacement lam.horsepower
#> Fold 1 9.71384 -0.17408 0.50000 0.00000 0.00000
#> Fold 2 9.21713 -0.31480 0.00000 0.00000 0.00000
#> Fold 3 9.61824 -0.19248 0.00000 0.00000 0.00000
#> Fold 4 8.69910 -0.25523 0.50000 0.00000 0.00000
#> Fold 5 9.14403 -0.14934 0.00000 0.00000 0.00000
#> Fold 6 9.63481 -0.16739 0.50000 0.00000 0.00000
#> Fold 7 9.98933 -0.36847 0.00000 0.00000 -0.15447
#> Fold 8 9.06301 -0.29721 0.00000 0.00000 0.00000
#> Fold 9 8.88315 -0.22684 0.00000 0.00000 0.00000
#> Fold 10 9.61727 -0.17086 0.00000 0.00000 0.00000
#> lam.weight lambda weight year71 year72 year73 year74
#> Fold 1 0.00000 0.00000 -0.74636 0.03764 -0.00327 -0.02477 0.05606
#> Fold 2 0.00000 0.00000 -0.47728 0.02173 -0.01488 -0.03770 0.04312
#> Fold 3 0.00000 0.00000 -0.72085 0.01128 -0.02569 -0.03872 0.05187
#> Fold 4 0.00000 0.00000 -0.53846 0.02153 -0.02922 -0.05181 0.04136
#> Fold 5 0.00000 0.00000 -0.69081 0.02531 -0.01062 -0.04625 0.05039
#> Fold 6 0.00000 0.00000 -0.74049 0.02456 0.00759 -0.03412 0.06266
#> Fold 7 0.00000 0.00000 -0.72843 0.02532 -0.01271 -0.04144 0.04568
#> Fold 8 0.00000 0.00000 -0.46392 0.02702 -0.02041 -0.05605 0.04437
#> Fold 9 0.00000 0.00000 -0.47136 0.00860 -0.03620 -0.04835 0.01906
#> Fold 10 0.00000 0.00000 -0.73550 0.02937 -0.00899 -0.03814 0.05408
#> year75 year76 year77 year78 year79 year80 year81 year82
#> Fold 1 0.07080 0.07250 0.14420 0.14281 0.23266 0.35127 0.25635 0.30546
#> Fold 2 0.04031 0.06718 0.13094 0.14917 0.21871 0.33192 0.26196 0.30943
#> Fold 3 0.03837 0.06399 0.11593 0.12601 0.20499 0.32821 0.24478 0.29204
#> Fold 4 0.04072 0.05537 0.12292 0.14083 0.22878 0.32947 0.25140 0.27248
#> Fold 5 0.05596 0.07044 0.13356 0.14724 0.24675 0.33331 0.26938 0.32594
#> Fold 6 0.06940 0.07769 0.14211 0.14647 0.23532 0.34761 0.26737 0.33062
#> Fold 7 0.03614 0.07385 0.12976 0.14040 0.23976 0.33998 0.27652 0.30659
#> Fold 8 0.06573 0.08135 0.13158 0.13987 0.23011 0.32880 0.25886 0.30538
#> Fold 9 0.03018 0.05846 0.10536 0.11722 0.20665 0.31533 0.23352 0.29375
#> Fold 10 0.04881 0.07862 0.14101 0.14313 0.23258 0.35649 0.26214 0.32421
#> acceleration displacement cylinders5.6 cylinders8 originEurope
#> Fold 1
#> Fold 2 -0.18909 -0.09197
#> Fold 3
#> Fold 4 -0.03484 -0.09080 -0.10909
#> Fold 5 0.06261
#> Fold 6
#> Fold 7
#> Fold 8 -0.17676 -0.10542
#> Fold 9 -0.14514 -0.13452
#> Fold 10
#> originJapan
#> Fold 1
#> Fold 2
#> Fold 3
#> Fold 4
#> Fold 5 0.04
#> Fold 6
#> Fold 7
#> Fold 8
#> Fold 9
#> Fold 10Here, as for selectTrans(), the predictors
and response arguments specify candidate variables for
transformation, and AIC=FALSE uses the BIC for model
selection. The starting model, m.auto, is the working model
fit to the Auto data. The CV criterion isn’t bias-adjusted
because median absolute error isn’t a mean of casewise error
components.
Some noteworthy points:
-
selectTransStepAIC()automatically computes CV cost criteria, here the median absolute error, on the untransformed response scale. - The estimate of the median absolute error that we obtain by
cross-validating the whole model-specification process is a little
larger than the median absolute error computed for the model we fit to
the
Autodata separately selecting transformations of the predictors and the response and then selecting predictors for the whole data set. - When we look at the transformations and predictors selected with
each of the 10 folds omitted (i.e., the output of
compareFolds()), we see that there is little uncertainty in choosing variable transformations (thelam.*s for the s andlambdafor in the output), but considerably more uncertainty in subsequently selecting predictors:horsepower,weight, andyearare always included among the selected predictors;accelerationanddisplacementare each included respectively in 4 and 3 of 10 selected models; andcylindersandoriginare each included in only 1 of 10 models. Recall that when we selected predictors for the full data, we obtained a model withhorsepower,weight,acceleration,year, andorigin.